Eric Z. Chen
DiffDenoise: Self-Supervised Medical Image Denoising with Conditional Diffusion Models
Mar 31, 2025Abstract:Many self-supervised denoising approaches have been proposed in recent years. However, these methods tend to overly smooth images, resulting in the loss of fine structures that are essential for medical applications. In this paper, we propose DiffDenoise, a powerful self-supervised denoising approach tailored for medical images, designed to preserve high-frequency details. Our approach comprises three stages. First, we train a diffusion model on noisy images, using the outputs of a pretrained Blind-Spot Network as conditioning inputs. Next, we introduce a novel stabilized reverse sampling technique, which generates clean images by averaging diffusion sampling outputs initialized with a pair of symmetric noises. Finally, we train a supervised denoising network using noisy images paired with the denoised outputs generated by the diffusion model. Our results demonstrate that DiffDenoise outperforms existing state-of-the-art methods in both synthetic and real-world medical image denoising tasks. We provide both a theoretical foundation and practical insights, demonstrating the method's effectiveness across various medical imaging modalities and anatomical structures.
Leveraging Diffusion Model and Image Foundation Model for Improved Correspondence Matching in Coronary Angiography
Mar 31, 2025Abstract:Accurate correspondence matching in coronary angiography images is crucial for reconstructing 3D coronary artery structures, which is essential for precise diagnosis and treatment planning of coronary artery disease (CAD). Traditional matching methods for natural images often fail to generalize to X-ray images due to inherent differences such as lack of texture, lower contrast, and overlapping structures, compounded by insufficient training data. To address these challenges, we propose a novel pipeline that generates realistic paired coronary angiography images using a diffusion model conditioned on 2D projections of 3D reconstructed meshes from Coronary Computed Tomography Angiography (CCTA), providing high-quality synthetic data for training. Additionally, we employ large-scale image foundation models to guide feature aggregation, enhancing correspondence matching accuracy by focusing on semantically relevant regions and keypoints. Our approach demonstrates superior matching performance on synthetic datasets and effectively generalizes to real-world datasets, offering a practical solution for this task. Furthermore, our work investigates the efficacy of different foundation models in correspondence matching, providing novel insights into leveraging advanced image foundation models for medical imaging applications.
Adapting Vision Foundation Models for Real-time Ultrasound Image Segmentation
Mar 31, 2025Abstract:We propose a novel approach that adapts hierarchical vision foundation models for real-time ultrasound image segmentation. Existing ultrasound segmentation methods often struggle with adaptability to new tasks, relying on costly manual annotations, while real-time approaches generally fail to match state-of-the-art performance. To overcome these limitations, we introduce an adaptive framework that leverages the vision foundation model Hiera to extract multi-scale features, interleaved with DINOv2 representations to enhance visual expressiveness. These enriched features are then decoded to produce precise and robust segmentation. We conduct extensive evaluations on six public datasets and one in-house dataset, covering both cardiac and thyroid ultrasound segmentation. Experiments show that our approach outperforms state-of-the-art methods across multiple datasets and excels with limited supervision, surpassing nnUNet by over 20\% on average in the 1\% and 10\% data settings. Our method achieves $\sim$77 FPS inference speed with TensorRT on a single GPU, enabling real-time clinical applications.
Label-Efficient Data Augmentation with Video Diffusion Models for Guidewire Segmentation in Cardiac Fluoroscopy
Dec 23, 2024Abstract:The accurate segmentation of guidewires in interventional cardiac fluoroscopy videos is crucial for computer-aided navigation tasks. Although deep learning methods have demonstrated high accuracy and robustness in wire segmentation, they require substantial annotated datasets for generalizability, underscoring the need for extensive labeled data to enhance model performance. To address this challenge, we propose the Segmentation-guided Frame-consistency Video Diffusion Model (SF-VD) to generate large collections of labeled fluoroscopy videos, augmenting the training data for wire segmentation networks. SF-VD leverages videos with limited annotations by independently modeling scene distribution and motion distribution. It first samples the scene distribution by generating 2D fluoroscopy images with wires positioned according to a specified input mask, and then samples the motion distribution by progressively generating subsequent frames, ensuring frame-to-frame coherence through a frame-consistency strategy. A segmentation-guided mechanism further refines the process by adjusting wire contrast, ensuring a diverse range of visibility in the synthesized image. Evaluation on a fluoroscopy dataset confirms the superior quality of the generated videos and shows significant improvements in guidewire segmentation.
SemiHVision: Enhancing Medical Multimodal Models with a Semi-Human Annotated Dataset and Fine-Tuned Instruction Generation
Oct 19, 2024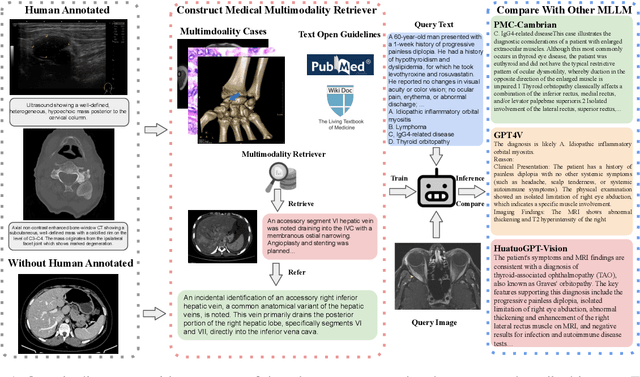
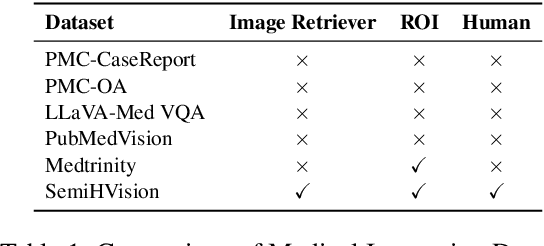
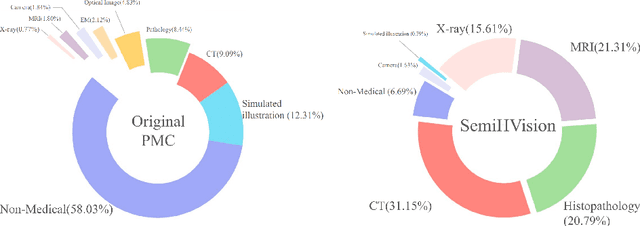
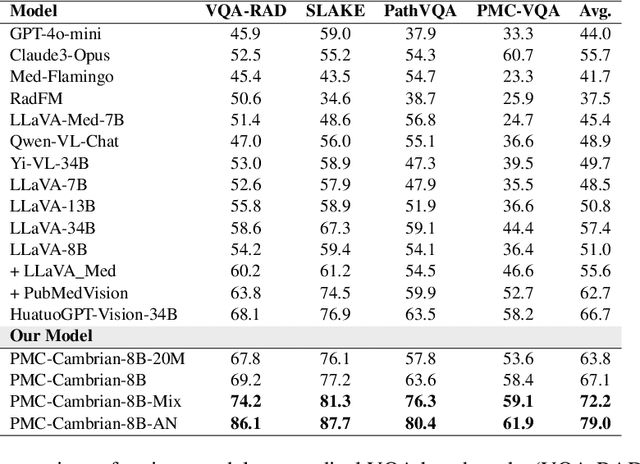
Abstract:Multimodal large language models (MLLMs) have made significant strides, yet they face challenges in the medical domain due to limited specialized knowledge. While recent medical MLLMs demonstrate strong performance in lab settings, they often struggle in real-world applications, highlighting a substantial gap between research and practice. In this paper, we seek to address this gap at various stages of the end-to-end learning pipeline, including data collection, model fine-tuning, and evaluation. At the data collection stage, we introduce SemiHVision, a dataset that combines human annotations with automated augmentation techniques to improve both medical knowledge representation and diagnostic reasoning. For model fine-tuning, we trained PMC-Cambrian-8B-AN over 2400 H100 GPU hours, resulting in performance that surpasses public medical models like HuatuoGPT-Vision-34B (79.0% vs. 66.7%) and private general models like Claude3-Opus (55.7%) on traditional benchmarks such as SLAKE and VQA-RAD. In the evaluation phase, we observed that traditional benchmarks cannot accurately reflect realistic clinical task capabilities. To overcome this limitation and provide more targeted guidance for model evaluation, we introduce the JAMA Clinical Challenge, a novel benchmark specifically designed to evaluate diagnostic reasoning. On this benchmark, PMC-Cambrian-AN achieves state-of-the-art performance with a GPT-4 score of 1.29, significantly outperforming HuatuoGPT-Vision-34B (1.13) and Claude3-Opus (1.17), demonstrating its superior diagnostic reasoning abilities.
Auxiliary Input in Training: Incorporating Catheter Features into Deep Learning Models for ECG-Free Dynamic Coronary Roadmapping
Aug 28, 2024



Abstract:Dynamic coronary roadmapping is a technology that overlays the vessel maps (the "roadmap") extracted from an offline image sequence of X-ray angiography onto a live stream of X-ray fluoroscopy in real-time. It aims to offer navigational guidance for interventional surgeries without the need for repeated contrast agent injections, thereby reducing the risks associated with radiation exposure and kidney failure. The precision of the roadmaps is contingent upon the accurate alignment of angiographic and fluoroscopic images based on their cardiac phases, as well as precise catheter tip tracking. The former ensures the selection of a roadmap that closely matches the vessel shape in the current frame, while the latter uses catheter tips as reference points to adjust for translational motion between the roadmap and the present vessel tree. Training deep learning models for both tasks is challenging and underexplored. However, incorporating catheter features into the models could offer substantial benefits, given humans heavily rely on catheters to complete the tasks. To this end, we introduce a simple but effective method, auxiliary input in training (AIT), and demonstrate that it enhances model performance across both tasks, outperforming baseline methods in knowledge incorporation and transfer learning.
Retrieval-augmented Few-shot Medical Image Segmentation with Foundation Models
Aug 16, 2024



Abstract:Medical image segmentation is crucial for clinical decision-making, but the scarcity of annotated data presents significant challenges. Few-shot segmentation (FSS) methods show promise but often require retraining on the target domain and struggle to generalize across different modalities. Similarly, adapting foundation models like the Segment Anything Model (SAM) for medical imaging has limitations, including the need for finetuning and domain-specific adaptation. To address these issues, we propose a novel method that adapts DINOv2 and Segment Anything Model 2 (SAM 2) for retrieval-augmented few-shot medical image segmentation. Our approach uses DINOv2's feature as query to retrieve similar samples from limited annotated data, which are then encoded as memories and stored in memory bank. With the memory attention mechanism of SAM 2, the model leverages these memories as conditions to generate accurate segmentation of the target image. We evaluated our framework on three medical image segmentation tasks, demonstrating superior performance and generalizability across various modalities without the need for any retraining or finetuning. Overall, this method offers a practical and effective solution for few-shot medical image segmentation and holds significant potential as a valuable annotation tool in clinical applications.
Spatiotemporal Diffusion Model with Paired Sampling for Accelerated Cardiac Cine MRI
Mar 13, 2024Abstract:Current deep learning reconstruction for accelerated cardiac cine MRI suffers from spatial and temporal blurring. We aim to improve image sharpness and motion delineation for cine MRI under high undersampling rates. A spatiotemporal diffusion enhancement model conditional on an existing deep learning reconstruction along with a novel paired sampling strategy was developed. The diffusion model provided sharper tissue boundaries and clearer motion than the original reconstruction in experts evaluation on clinical data. The innovative paired sampling strategy substantially reduced artificial noises in the generative results.
Clinically Feasible Diffusion Reconstruction for Highly-Accelerated Cardiac Cine MRI
Mar 13, 2024Abstract:The currently limited quality of accelerated cardiac cine reconstruction may potentially be improved by the emerging diffusion models, but the clinically unacceptable long processing time poses a challenge. We aim to develop a clinically feasible diffusion-model-based reconstruction pipeline to improve the image quality of cine MRI. A multi-in multi-out diffusion enhancement model together with fast inference strategies were developed to be used in conjunction with a reconstruction model. The diffusion reconstruction reduced spatial and temporal blurring in prospectively undersampled clinical data, as validated by experts inspection. The 1.5s per video processing time enabled the approach to be applied in clinical scenarios.
Federated Data Model
Mar 13, 2024Abstract:In artificial intelligence (AI), especially deep learning, data diversity and volume play a pivotal role in model development. However, training a robust deep learning model often faces challenges due to data privacy, regulations, and the difficulty of sharing data between different locations, especially for medical applications. To address this, we developed a method called the Federated Data Model (FDM). This method uses diffusion models to learn the characteristics of data at one site and then creates synthetic data that can be used at another site without sharing the actual data. We tested this approach with a medical image segmentation task, focusing on cardiac magnetic resonance images from different hospitals. Our results show that models trained with this method perform well both on the data they were originally trained on and on data from other sites. This approach offers a promising way to train accurate and privacy-respecting AI models across different locations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge