Hieu Tran
PRIME: Planning and Retrieval-Integrated Memory for Enhanced Reasoning
Sep 26, 2025Abstract:Inspired by the dual-process theory of human cognition from \textit{Thinking, Fast and Slow}, we introduce \textbf{PRIME} (Planning and Retrieval-Integrated Memory for Enhanced Reasoning), a multi-agent reasoning framework that dynamically integrates \textbf{System 1} (fast, intuitive thinking) and \textbf{System 2} (slow, deliberate thinking). PRIME first employs a Quick Thinking Agent (System 1) to generate a rapid answer; if uncertainty is detected, it then triggers a structured System 2 reasoning pipeline composed of specialized agents for \textit{planning}, \textit{hypothesis generation}, \textit{retrieval}, \textit{information integration}, and \textit{decision-making}. This multi-agent design faithfully mimics human cognitive processes and enhances both efficiency and accuracy. Experimental results with LLaMA 3 models demonstrate that PRIME enables open-source LLMs to perform competitively with state-of-the-art closed-source models like GPT-4 and GPT-4o on benchmarks requiring multi-hop and knowledge-grounded reasoning. This research establishes PRIME as a scalable solution for improving LLMs in domains requiring complex, knowledge-intensive reasoning.
A Robot That Listens: Enhancing Self-Disclosure and Engagement Through Sentiment-based Backchannels and Active Listening
Sep 09, 2025Abstract:As social robots get more deeply integrated intoour everyday lives, they will be expected to engage in meaningful conversations and exhibit socio-emotionally intelligent listening behaviors when interacting with people. Active listening and backchanneling could be one way to enhance robots' communicative capabilities and enhance their effectiveness in eliciting deeper self-disclosure, providing a sense of empathy,and forming positive rapport and relationships with people.Thus, we developed an LLM-powered social robot that can exhibit contextually appropriate sentiment-based backchannelingand active listening behaviors (active listening+backchanneling) and compared its efficacy in eliciting people's self-disclosurein comparison to robots that do not exhibit any of these listening behaviors (control) and a robot that only exhibitsbackchanneling behavior (backchanneling-only). Through ourexperimental study with sixty-five participants, we found theparticipants who conversed with the active listening robot per-ceived the interactions more positively, in which they exhibited the highest self-disclosures, and reported the strongest senseof being listened to. The results of our study suggest that the implementation of active listening behaviors in social robotshas the potential to improve human-robot communication andcould further contribute to the building of deeper human-robot relationships and rapport.
MedReadCtrl: Personalizing medical text generation with readability-controlled instruction learning
Jul 10, 2025Abstract:Generative AI has demonstrated strong potential in healthcare, from clinical decision support to patient-facing chatbots that improve outcomes. A critical challenge for deployment is effective human-AI communication, where content must be both personalized and understandable. We introduce MedReadCtrl, a readability-controlled instruction tuning framework that enables LLMs to adjust output complexity without compromising meaning. Evaluations of nine datasets and three tasks across medical and general domains show that MedReadCtrl achieves significantly lower readability instruction-following errors than GPT-4 (e.g., 1.39 vs. 1.59 on ReadMe, p<0.001) and delivers substantial gains on unseen clinical tasks (e.g., +14.7 ROUGE-L, +6.18 SARI on MTSamples). Experts consistently preferred MedReadCtrl (71.7% vs. 23.3%), especially at low literacy levels. These gains reflect MedReadCtrl's ability to restructure clinical content into accessible, readability-aligned language while preserving medical intent, offering a scalable solution to support patient education and expand equitable access to AI-enabled care.
VietMix: A Naturally Occurring Vietnamese-English Code-Mixed Corpus with Iterative Augmentation for Machine Translation
May 30, 2025Abstract:Machine translation systems fail when processing code-mixed inputs for low-resource languages. We address this challenge by curating VietMix, a parallel corpus of naturally occurring code-mixed Vietnamese text paired with expert English translations. Augmenting this resource, we developed a complementary synthetic data generation pipeline. This pipeline incorporates filtering mechanisms to ensure syntactic plausibility and pragmatic appropriateness in code-mixing patterns. Experimental validation shows our naturalistic and complementary synthetic data boost models' performance, measured by translation quality estimation scores, of up to 71.84 on COMETkiwi and 81.77 on XCOMET. Triangulating positive results with LLM-based assessments, augmented models are favored over seed fine-tuned counterparts in approximately 49% of judgments (54-56% excluding ties). VietMix and our augmentation methodology advance ecological validity in neural MT evaluations and establish a framework for addressing code-mixed translation challenges across other low-resource pairs.
Enhancing LLMs for Identifying and Prioritizing Important Medical Jargons from Electronic Health Record Notes Utilizing Data Augmentation
Feb 25, 2025Abstract:OpenNotes enables patients to access EHR notes, but medical jargon can hinder comprehension. To improve understanding, we evaluated closed- and open-source LLMs for extracting and prioritizing key medical terms using prompting, fine-tuning, and data augmentation. We assessed LLMs on 106 expert-annotated EHR notes, experimenting with (i) general vs. structured prompts, (ii) zero-shot vs. few-shot prompting, (iii) fine-tuning, and (iv) data augmentation. To enhance open-source models in low-resource settings, we used ChatGPT for data augmentation and applied ranking techniques. We incrementally increased the augmented dataset size (10 to 10,000) and conducted 5-fold cross-validation, reporting F1 score and Mean Reciprocal Rank (MRR). Our result show that fine-tuning and data augmentation improved performance over other strategies. GPT-4 Turbo achieved the highest F1 (0.433), while Mistral7B with data augmentation had the highest MRR (0.746). Open-source models, when fine-tuned or augmented, outperformed closed-source models. Notably, the best F1 and MRR scores did not always align. Few-shot prompting outperformed zero-shot in vanilla models, and structured prompts yielded different preferences across models. Fine-tuning improved zero-shot performance but sometimes degraded few-shot performance. Data augmentation performed comparably or better than other methods. Our evaluation highlights the effectiveness of prompting, fine-tuning, and data augmentation in improving model performance for medical jargon extraction in low-resource scenarios.
RARE: Retrieval-Augmented Reasoning Enhancement for Large Language Models
Dec 05, 2024Abstract:This work introduces RARE (Retrieval-Augmented Reasoning Enhancement), a versatile extension to the mutual reasoning framework (rStar), aimed at enhancing reasoning accuracy and factual integrity across large language models (LLMs) for complex, knowledge-intensive tasks such as commonsense and medical reasoning. RARE incorporates two innovative actions within the Monte Carlo Tree Search (MCTS) framework: A6, which generates search queries based on the initial problem statement, performs information retrieval using those queries, and augments reasoning with the retrieved data to formulate the final answer; and A7, which leverages information retrieval specifically for generated sub-questions and re-answers these sub-questions with the relevant contextual information. Additionally, a Retrieval-Augmented Factuality Scorer is proposed to replace the original discriminator, prioritizing reasoning paths that meet high standards of factuality. Experimental results with LLaMA 3.1 show that RARE enables open-source LLMs to achieve competitive performance with top open-source models like GPT-4 and GPT-4o. This research establishes RARE as a scalable solution for improving LLMs in domains where logical coherence and factual integrity are critical.
LEAF: Learning and Evaluation Augmented by Fact-Checking to Improve Factualness in Large Language Models
Oct 31, 2024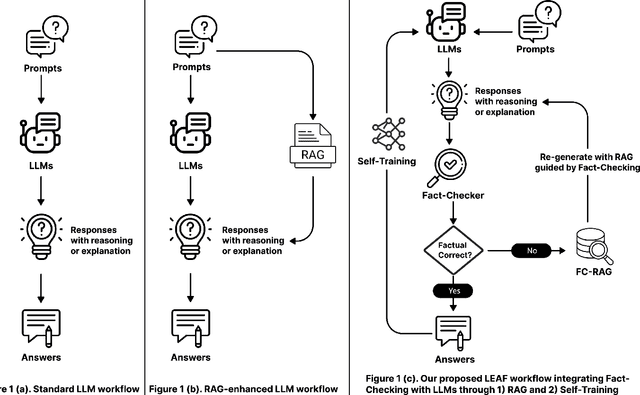

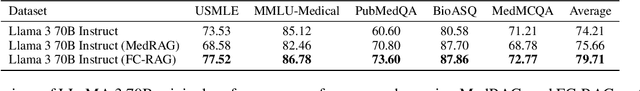

Abstract:Large language models (LLMs) have shown remarkable capabilities in various natural language processing tasks, yet they often struggle with maintaining factual accuracy, particularly in knowledge-intensive domains like healthcare. This study introduces LEAF: Learning and Evaluation Augmented by Fact-Checking, a novel approach designed to enhance the factual reliability of LLMs, with a focus on medical question answering (QA). LEAF utilizes a dual strategy to enhance the factual accuracy of responses from models such as Llama 3 70B Instruct and Llama 3 8B Instruct. The first strategy, Fact-Check-Then-RAG, improves Retrieval-Augmented Generation (RAG) by incorporating fact-checking results to guide the retrieval process without updating model parameters. The second strategy, Learning from Fact-Checks via Self-Training, involves supervised fine-tuning (SFT) on fact-checked responses or applying Simple Preference Optimization (SimPO) with fact-checking as a ranking mechanism, both updating LLM parameters from supervision. These findings suggest that integrating fact-checked responses whether through RAG enhancement or self-training enhances the reliability and factual correctness of LLM outputs, offering a promising solution for applications where information accuracy is crucial.
SemiHVision: Enhancing Medical Multimodal Models with a Semi-Human Annotated Dataset and Fine-Tuned Instruction Generation
Oct 19, 2024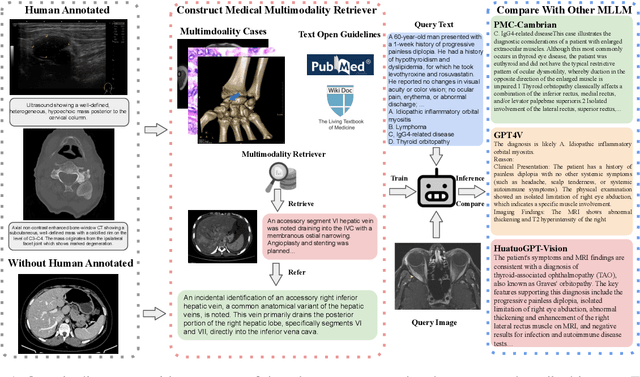
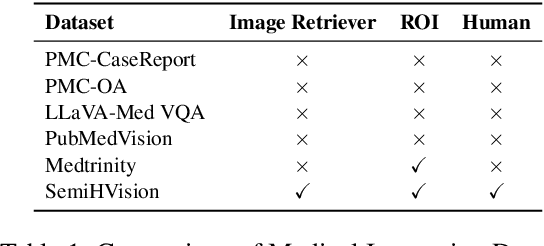
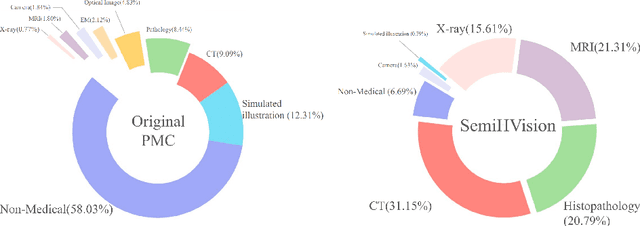
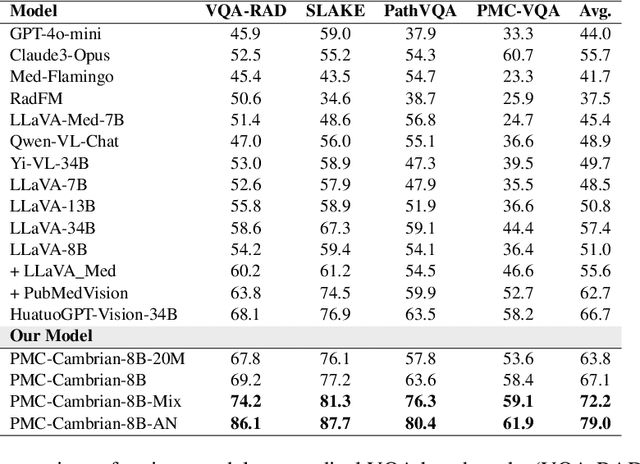
Abstract:Multimodal large language models (MLLMs) have made significant strides, yet they face challenges in the medical domain due to limited specialized knowledge. While recent medical MLLMs demonstrate strong performance in lab settings, they often struggle in real-world applications, highlighting a substantial gap between research and practice. In this paper, we seek to address this gap at various stages of the end-to-end learning pipeline, including data collection, model fine-tuning, and evaluation. At the data collection stage, we introduce SemiHVision, a dataset that combines human annotations with automated augmentation techniques to improve both medical knowledge representation and diagnostic reasoning. For model fine-tuning, we trained PMC-Cambrian-8B-AN over 2400 H100 GPU hours, resulting in performance that surpasses public medical models like HuatuoGPT-Vision-34B (79.0% vs. 66.7%) and private general models like Claude3-Opus (55.7%) on traditional benchmarks such as SLAKE and VQA-RAD. In the evaluation phase, we observed that traditional benchmarks cannot accurately reflect realistic clinical task capabilities. To overcome this limitation and provide more targeted guidance for model evaluation, we introduce the JAMA Clinical Challenge, a novel benchmark specifically designed to evaluate diagnostic reasoning. On this benchmark, PMC-Cambrian-AN achieves state-of-the-art performance with a GPT-4 score of 1.29, significantly outperforming HuatuoGPT-Vision-34B (1.13) and Claude3-Opus (1.17), demonstrating its superior diagnostic reasoning abilities.
ReadCtrl: Personalizing text generation with readability-controlled instruction learning
Jun 13, 2024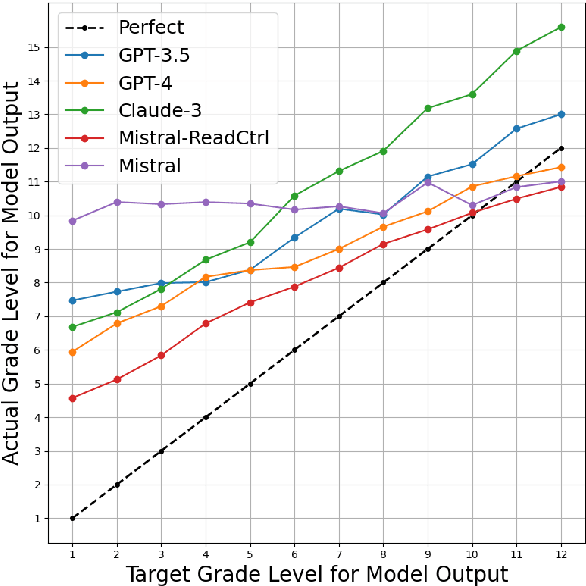
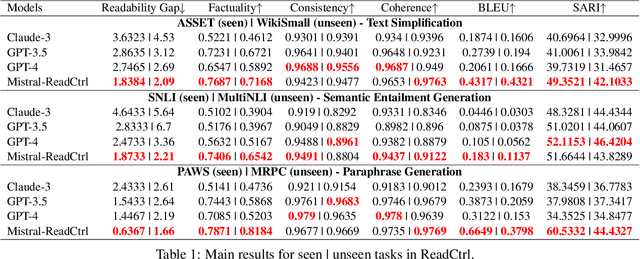
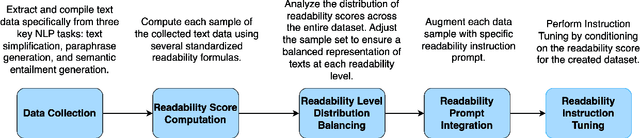
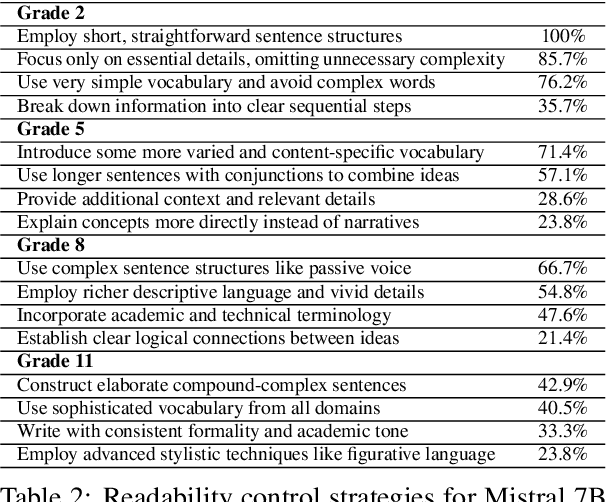
Abstract:Content generation conditioning on users's readability is an important application for personalization. In an era of large language models (LLMs), readability-controlled text generation based on LLMs has become increasingly important. This paper introduces a novel methodology called "Readability-Controlled Instruction Learning (ReadCtrl)," which aims to instruction-tune LLMs to tailor users' readability levels. Unlike the traditional methods, which primarily focused on categorical readability adjustments typically classified as high, medium, and low or expert and layperson levels with limited success, ReadCtrl introduces a dynamic framework that enables LLMs to generate content at various (near continuous level) complexity levels, thereby enhancing their versatility across different applications. Our results show that the ReadCtrl-Mistral-7B models significantly outperformed strong baseline models such as GPT-4 and Claude-3, with a win rate of 52.1%:35.7% against GPT-4 in human evaluations. Furthermore, Read-Ctrl has shown significant improvements in automatic evaluations, as evidenced by better readability metrics (e.g., FOG, FKGL) and generation quality metrics (e.g., BLEU, SARI, SummaC-Factuality, UniEval-Consistency and Coherence). These results underscore Read-Ctrl's effectiveness and tenacity in producing high-quality, contextually appropriate outputs that closely align with targeted readability levels, marking a significant advancement in personalized content generation using LLMs.
Distillation Contrastive Decoding: Improving LLMs Reasoning with Contrastive Decoding and Distillation
Feb 21, 2024Abstract:We propose a straightforward approach called Distillation Contrastive Decoding (DCD) to enhance the reasoning capabilities of Large Language Models (LLMs) during inference. In contrast to previous approaches that relied on smaller amateur models or analysis of hidden state differences, DCD employs Contrastive Chain-of-thought Prompting and advanced distillation techniques, including Dropout and Quantization. This approach effectively addresses the limitations of Contrastive Decoding (CD), which typically requires both an expert and an amateur model, thus increasing computational resource demands. By integrating contrastive prompts with distillation, DCD obviates the need for an amateur model and reduces memory usage. Our evaluations demonstrate that DCD significantly enhances LLM performance across a range of reasoning benchmarks, surpassing both CD and existing methods in the GSM8K and StrategyQA datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge