Weijing Huang
LEAF: Learning and Evaluation Augmented by Fact-Checking to Improve Factualness in Large Language Models
Oct 31, 2024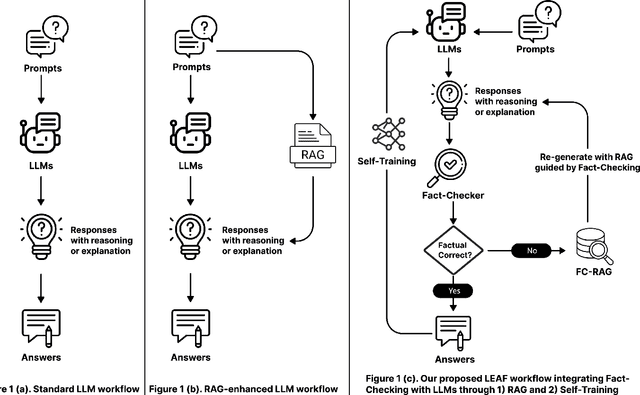

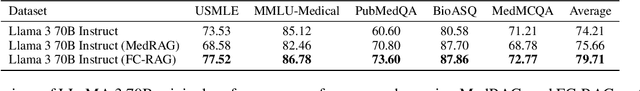

Abstract:Large language models (LLMs) have shown remarkable capabilities in various natural language processing tasks, yet they often struggle with maintaining factual accuracy, particularly in knowledge-intensive domains like healthcare. This study introduces LEAF: Learning and Evaluation Augmented by Fact-Checking, a novel approach designed to enhance the factual reliability of LLMs, with a focus on medical question answering (QA). LEAF utilizes a dual strategy to enhance the factual accuracy of responses from models such as Llama 3 70B Instruct and Llama 3 8B Instruct. The first strategy, Fact-Check-Then-RAG, improves Retrieval-Augmented Generation (RAG) by incorporating fact-checking results to guide the retrieval process without updating model parameters. The second strategy, Learning from Fact-Checks via Self-Training, involves supervised fine-tuning (SFT) on fact-checked responses or applying Simple Preference Optimization (SimPO) with fact-checking as a ranking mechanism, both updating LLM parameters from supervision. These findings suggest that integrating fact-checked responses whether through RAG enhancement or self-training enhances the reliability and factual correctness of LLM outputs, offering a promising solution for applications where information accuracy is crucial.
SemiHVision: Enhancing Medical Multimodal Models with a Semi-Human Annotated Dataset and Fine-Tuned Instruction Generation
Oct 19, 2024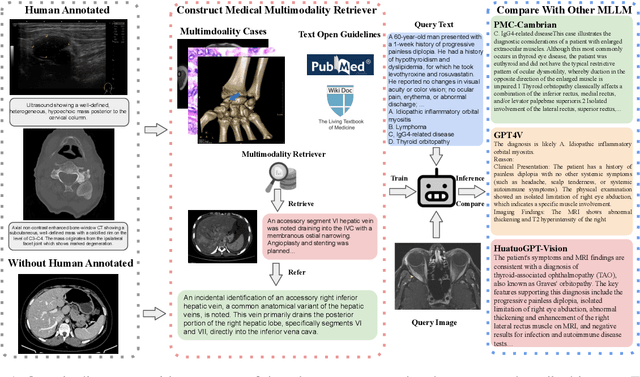
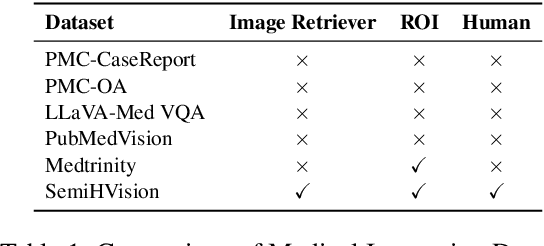
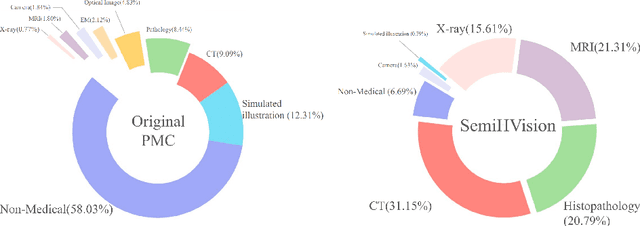
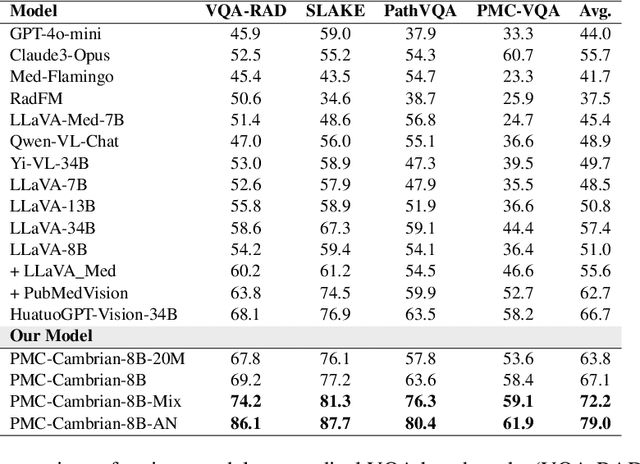
Abstract:Multimodal large language models (MLLMs) have made significant strides, yet they face challenges in the medical domain due to limited specialized knowledge. While recent medical MLLMs demonstrate strong performance in lab settings, they often struggle in real-world applications, highlighting a substantial gap between research and practice. In this paper, we seek to address this gap at various stages of the end-to-end learning pipeline, including data collection, model fine-tuning, and evaluation. At the data collection stage, we introduce SemiHVision, a dataset that combines human annotations with automated augmentation techniques to improve both medical knowledge representation and diagnostic reasoning. For model fine-tuning, we trained PMC-Cambrian-8B-AN over 2400 H100 GPU hours, resulting in performance that surpasses public medical models like HuatuoGPT-Vision-34B (79.0% vs. 66.7%) and private general models like Claude3-Opus (55.7%) on traditional benchmarks such as SLAKE and VQA-RAD. In the evaluation phase, we observed that traditional benchmarks cannot accurately reflect realistic clinical task capabilities. To overcome this limitation and provide more targeted guidance for model evaluation, we introduce the JAMA Clinical Challenge, a novel benchmark specifically designed to evaluate diagnostic reasoning. On this benchmark, PMC-Cambrian-AN achieves state-of-the-art performance with a GPT-4 score of 1.29, significantly outperforming HuatuoGPT-Vision-34B (1.13) and Claude3-Opus (1.17), demonstrating its superior diagnostic reasoning abilities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge