Basar Demir
On The Robustness of Foundational 3D Medical Image Segmentation Models Against Imprecise Visual Prompts
Jan 23, 2026Abstract:While 3D foundational models have shown promise for promptable segmentation of medical volumes, their robustness to imprecise prompts remains under-explored. In this work, we aim to address this gap by systematically studying the effect of various controlled perturbations of dense visual prompts, that closely mimic real-world imprecision. By conducting experiments with two recent foundational models on a multi-organ abdominal segmentation task, we reveal several facets of promptable medical segmentation, especially pertaining to reliance on visual shape and spatial cues, and the extent of resilience of models towards certain perturbations. Codes are available at: https://github.com/ucsdbiag/Prompt-Robustness-MedSegFMs
DiffDenoise: Self-Supervised Medical Image Denoising with Conditional Diffusion Models
Mar 31, 2025Abstract:Many self-supervised denoising approaches have been proposed in recent years. However, these methods tend to overly smooth images, resulting in the loss of fine structures that are essential for medical applications. In this paper, we propose DiffDenoise, a powerful self-supervised denoising approach tailored for medical images, designed to preserve high-frequency details. Our approach comprises three stages. First, we train a diffusion model on noisy images, using the outputs of a pretrained Blind-Spot Network as conditioning inputs. Next, we introduce a novel stabilized reverse sampling technique, which generates clean images by averaging diffusion sampling outputs initialized with a pair of symmetric noises. Finally, we train a supervised denoising network using noisy images paired with the denoised outputs generated by the diffusion model. Our results demonstrate that DiffDenoise outperforms existing state-of-the-art methods in both synthetic and real-world medical image denoising tasks. We provide both a theoretical foundation and practical insights, demonstrating the method's effectiveness across various medical imaging modalities and anatomical structures.
Zero-shot Domain Generalization of Foundational Models for 3D Medical Image Segmentation: An Experimental Study
Mar 28, 2025Abstract:Domain shift, caused by variations in imaging modalities and acquisition protocols, limits model generalization in medical image segmentation. While foundation models (FMs) trained on diverse large-scale data hold promise for zero-shot generalization, their application to volumetric medical data remains underexplored. In this study, we examine their ability towards domain generalization (DG), by conducting a comprehensive experimental study encompassing 6 medical segmentation FMs and 12 public datasets spanning multiple modalities and anatomies. Our findings reveal the potential of promptable FMs in bridging the domain gap via smart prompting techniques. Additionally, by probing into multiple facets of zero-shot DG, we offer valuable insights into the viability of FMs for DG and identify promising avenues for future research.
Downstream Analysis of Foundational Medical Vision Models for Disease Progression
Mar 21, 2025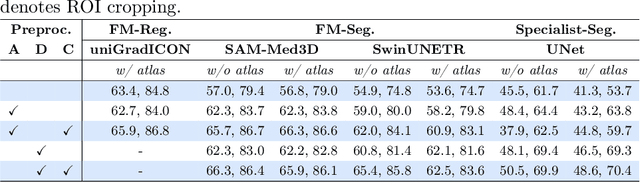
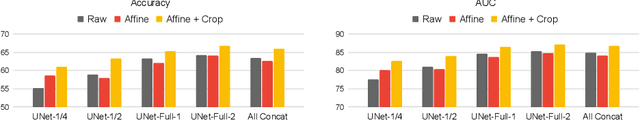
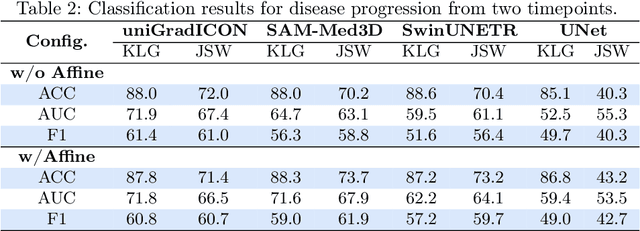
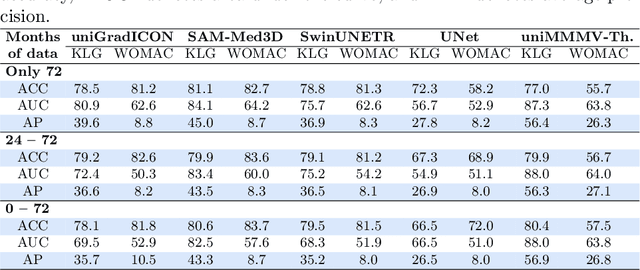
Abstract:Medical vision foundational models are used for a wide variety of tasks, including medical image segmentation and registration. This work evaluates the ability of these models to predict disease progression using a simple linear probe. We hypothesize that intermediate layer features of segmentation models capture structural information, while those of registration models encode knowledge of change over time. Beyond demonstrating that these features are useful for disease progression prediction, we also show that registration model features do not require spatially aligned input images. However, for segmentation models, spatial alignment is essential for optimal performance. Our findings highlight the importance of spatial alignment and the utility of foundation model features for image registration.
multiGradICON: A Foundation Model for Multimodal Medical Image Registration
Aug 01, 2024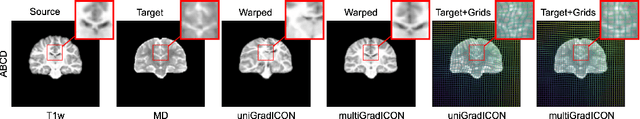
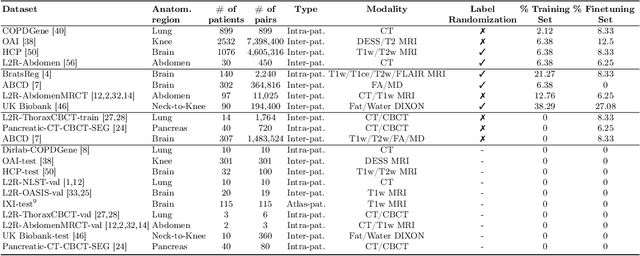
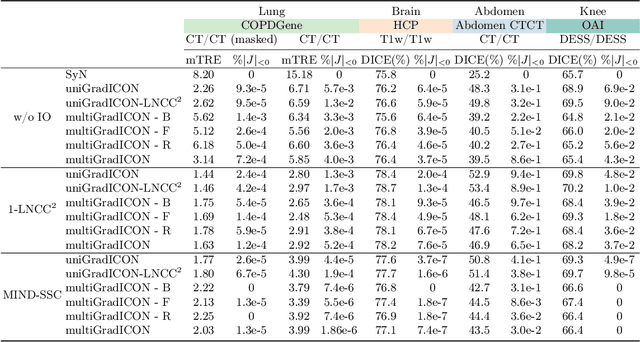
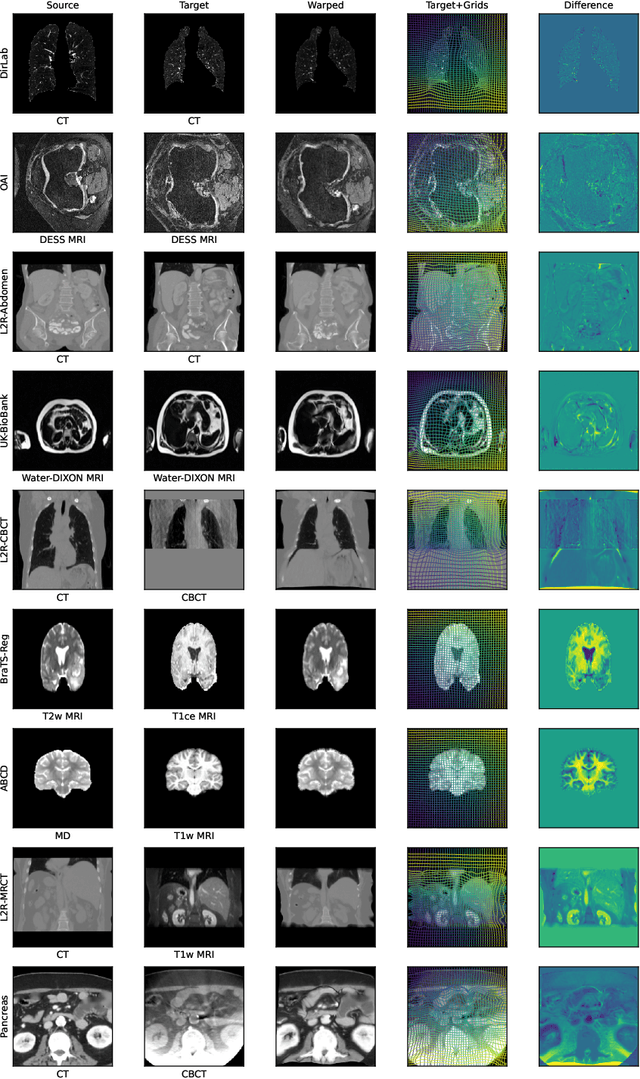
Abstract:Modern medical image registration approaches predict deformations using deep networks. These approaches achieve state-of-the-art (SOTA) registration accuracy and are generally fast. However, deep learning (DL) approaches are, in contrast to conventional non-deep-learning-based approaches, anatomy-specific. Recently, a universal deep registration approach, uniGradICON, has been proposed. However, uniGradICON focuses on monomodal image registration. In this work, we therefore develop multiGradICON as a first step towards universal *multimodal* medical image registration. Specifically, we show that 1) we can train a DL registration model that is suitable for monomodal *and* multimodal registration; 2) loss function randomization can increase multimodal registration accuracy; and 3) training a model with multimodal data helps multimodal generalization. Our code and the multiGradICON model are available at https://github.com/uncbiag/uniGradICON.
Inter-Domain Alignment for Predicting High-Resolution Brain Networks Using Teacher-Student Learning
Oct 06, 2021


Abstract:Accurate and automated super-resolution image synthesis is highly desired since it has the great potential to circumvent the need for acquiring high-cost medical scans and a time-consuming preprocessing pipeline of neuroimaging data. However, existing deep learning frameworks are solely designed to predict high-resolution (HR) image from a low-resolution (LR) one, which limits their generalization ability to brain graphs (i.e., connectomes). A small body of works has focused on superresolving brain graphs where the goal is to predict a HR graph from a single LR graph. Although promising, existing works mainly focus on superresolving graphs belonging to the same domain (e.g., functional), overlooking the domain fracture existing between multimodal brain data distributions (e.g., morphological and structural). To this aim, we propose a novel inter-domain adaptation framework namely, Learn to SuperResolve Brain Graphs with Knowledge Distillation Network (L2S-KDnet), which adopts a teacher-student paradigm to superresolve brain graphs. Our teacher network is a graph encoder-decoder that firstly learns the LR brain graph embeddings, and secondly learns how to align the resulting latent representations to the HR ground truth data distribution using an adversarial regularization. Ultimately, it decodes the HR graphs from the aligned embeddings. Next, our student network learns the knowledge of the aligned brain graphs as well as the topological structure of the predicted HR graphs transferred from the teacher. We further leverage the decoder of the teacher to optimize the student network. L2S-KDnet presents the first TS architecture tailored for brain graph super-resolution synthesis that is based on inter-domain alignment. Our experimental results demonstrate substantial performance gains over benchmark methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge