Islem Rekik
Rethinking Graph Super-resolution: Dual Frameworks for Topological Fidelity
Nov 12, 2025



Abstract:Graph super-resolution, the task of inferring high-resolution (HR) graphs from low-resolution (LR) counterparts, is an underexplored yet crucial research direction that circumvents the need for costly data acquisition. This makes it especially desirable for resource-constrained fields such as the medical domain. While recent GNN-based approaches show promise, they suffer from two key limitations: (1) matrix-based node super-resolution that disregards graph structure and lacks permutation invariance; and (2) reliance on node representations to infer edge weights, which limits scalability and expressivity. In this work, we propose two GNN-agnostic frameworks to address these issues. First, Bi-SR introduces a bipartite graph connecting LR and HR nodes to enable structure-aware node super-resolution that preserves topology and permutation invariance. Second, DEFEND learns edge representations by mapping HR edges to nodes of a dual graph, allowing edge inference via standard node-based GNNs. We evaluate both frameworks on a real-world brain connectome dataset, where they achieve state-of-the-art performance across seven topological measures. To support generalization, we introduce twelve new simulated datasets that capture diverse topologies and LR-HR relationships. These enable comprehensive benchmarking of graph super-resolution methods.
UnifiedFL: A Dynamic Unified Learning Framework for Equitable Federation
Oct 30, 2025Abstract:Federated learning (FL) has emerged as a key paradigm for collaborative model training across multiple clients without sharing raw data, enabling privacy-preserving applications in areas such as radiology and pathology. However, works on collaborative training across clients with fundamentally different neural architectures and non-identically distributed datasets remain scarce. Existing FL frameworks face several limitations. Despite claiming to support architectural heterogeneity, most recent FL methods only tolerate variants within a single model family (e.g., shallower, deeper, or wider CNNs), still presuming a shared global architecture and failing to accommodate federations where clients deploy fundamentally different network types (e.g., CNNs, GNNs, MLPs). Moreover, existing approaches often address only statistical heterogeneity while overlooking the domain-fracture problem, where each client's data distribution differs markedly from that faced at testing time, undermining model generalizability. When clients use different architectures, have non-identically distributed data, and encounter distinct test domains, current methods perform poorly. To address these challenges, we propose UnifiedFL, a dynamic federated learning framework that represents heterogeneous local networks as nodes and edges in a directed model graph optimized by a shared graph neural network (GNN). UnifiedFL introduces (i) a common GNN to parameterize all architectures, (ii) distance-driven clustering via Euclidean distances between clients' parameters, and (iii) a two-tier aggregation policy balancing convergence and diversity. Experiments on MedMNIST classification and hippocampus segmentation benchmarks demonstrate UnifiedFL's superior performance. Code and data: https://github.com/basiralab/UnifiedFL
DeltaGNN: Graph Neural Network with Information Flow Control
Jan 10, 2025Abstract:Graph Neural Networks (GNNs) are popular deep learning models designed to process graph-structured data through recursive neighborhood aggregations in the message passing process. When applied to semi-supervised node classification, the message-passing enables GNNs to understand short-range spatial interactions, but also causes them to suffer from over-smoothing and over-squashing. These challenges hinder model expressiveness and prevent the use of deeper models to capture long-range node interactions (LRIs) within the graph. Popular solutions for LRIs detection are either too expensive to process large graphs due to high time complexity or fail to generalize across diverse graph structures. To address these limitations, we propose a mechanism called \emph{information flow control}, which leverages a novel connectivity measure, called \emph{information flow score}, to address over-smoothing and over-squashing with linear computational overhead, supported by theoretical evidence. Finally, to prove the efficacy of our methodology we design DeltaGNN, the first scalable and generalizable approach for detecting long-range and short-range interactions. We benchmark our model across 10 real-world datasets, including graphs with varying sizes, topologies, densities, and homophilic ratios, showing superior performance with limited computational complexity. The implementation of the proposed methods are publicly available at https://github.com/basiralab/DeltaGNN.
Strongly Topology-preserving GNNs for Brain Graph Super-resolution
Nov 01, 2024Abstract:Brain graph super-resolution (SR) is an under-explored yet highly relevant task in network neuroscience. It circumvents the need for costly and time-consuming medical imaging data collection, preparation, and processing. Current SR methods leverage graph neural networks (GNNs) thanks to their ability to natively handle graph-structured datasets. However, most GNNs perform node feature learning, which presents two significant limitations: (1) they require computationally expensive methods to learn complex node features capable of inferring connectivity strength or edge features, which do not scale to larger graphs; and (2) computations in the node space fail to adequately capture higher-order brain topologies such as cliques and hubs. However, numerous studies have shown that brain graph topology is crucial in identifying the onset and presence of various neurodegenerative disorders like Alzheimer and Parkinson. Motivated by these challenges and applications, we propose our STP-GSR framework. It is the first graph SR architecture to perform representation learning in higher-order topological space. Specifically, using the primal-dual graph formulation from graph theory, we develop an efficient mapping from the edge space of our low-resolution (LR) brain graphs to the node space of a high-resolution (HR) dual graph. This approach ensures that node-level computations on this dual graph correspond naturally to edge-level learning on our HR brain graphs, thereby enforcing strong topological consistency within our framework. Additionally, our framework is GNN layer agnostic and can easily learn from smaller, scalable GNNs, reducing computational requirements. We comprehensively benchmark our framework across seven key topological measures and observe that it significantly outperforms the previous state-of-the-art methods and baselines.
Metadata-Driven Federated Learning of Connectional Brain Templates in Non-IID Multi-Domain Scenarios
Mar 14, 2024



Abstract:A connectional brain template (CBT) is a holistic representation of a population of multi-view brain connectivity graphs, encoding shared patterns and normalizing typical variations across individuals. The federation of CBT learning allows for an inclusive estimation of the representative center of multi-domain brain connectivity datasets in a fully data-preserving manner. However, existing methods overlook the non-independent and identically distributed (non-IDD) issue stemming from multidomain brain connectivity heterogeneity, in which data domains are drawn from different hospitals and imaging modalities. To overcome this limitation, we unprecedentedly propose a metadata-driven federated learning framework, called MetaFedCBT, for cross-domain CBT learning. Given the data drawn from a specific domain (i.e., hospital), our model aims to learn metadata in a fully supervised manner by introducing a local client-based regressor network. The generated meta-data is forced to meet the statistical attributes (e.g., mean) of other domains, while preserving their privacy. Our supervised meta-data generation approach boosts the unsupervised learning of a more centered, representative, and holistic CBT of a particular brain state across diverse domains. As the federated learning progresses over multiple rounds, the learned metadata and associated generated connectivities are continuously updated to better approximate the target domain information. MetaFedCBT overcomes the non-IID issue of existing methods by generating informative brain connectivities for privacy-preserving holistic CBT learning with guidance using metadata. Extensive experiments on multi-view morphological brain networks of normal and patient subjects demonstrate that our MetaFedCBT is a superior federated CBT learning model and significantly advances the state-of-the-art performance.
Predicting Infant Brain Connectivity with Federated Multi-Trajectory GNNs using Scarce Data
Jan 08, 2024



Abstract:The understanding of the convoluted evolution of infant brain networks during the first postnatal year is pivotal for identifying the dynamics of early brain connectivity development. Existing deep learning solutions suffer from three major limitations. First, they cannot generalize to multi-trajectory prediction tasks, where each graph trajectory corresponds to a particular imaging modality or connectivity type (e.g., T1-w MRI). Second, existing models require extensive training datasets to achieve satisfactory performance which are often challenging to obtain. Third, they do not efficiently utilize incomplete time series data. To address these limitations, we introduce FedGmTE-Net++, a federated graph-based multi-trajectory evolution network. Using the power of federation, we aggregate local learnings among diverse hospitals with limited datasets. As a result, we enhance the performance of each hospital's local generative model, while preserving data privacy. The three key innovations of FedGmTE-Net++ are: (i) presenting the first federated learning framework specifically designed for brain multi-trajectory evolution prediction in a data-scarce environment, (ii) incorporating an auxiliary regularizer in the local objective function to exploit all the longitudinal brain connectivity within the evolution trajectory and maximize data utilization, (iii) introducing a two-step imputation process, comprising a preliminary KNN-based precompletion followed by an imputation refinement step that employs regressors to improve similarity scores and refine imputations. Our comprehensive experimental results showed the outperformance of FedGmTE-Net++ in brain multi-trajectory prediction from a single baseline graph in comparison with benchmark methods.
Replica Tree-based Federated Learning using Limited Data
Dec 28, 2023



Abstract:Learning from limited data has been extensively studied in machine learning, considering that deep neural networks achieve optimal performance when trained using a large amount of samples. Although various strategies have been proposed for centralized training, the topic of federated learning with small datasets remains largely unexplored. Moreover, in realistic scenarios, such as settings where medical institutions are involved, the number of participating clients is also constrained. In this work, we propose a novel federated learning framework, named RepTreeFL. At the core of the solution is the concept of a replica, where we replicate each participating client by copying its model architecture and perturbing its local data distribution. Our approach enables learning from limited data and a small number of clients by aggregating a larger number of models with diverse data distributions. Furthermore, we leverage the hierarchical structure of the client network (both original and virtual), alongside the model diversity across replicas, and introduce a diversity-based tree aggregation, where replicas are combined in a tree-like manner and the aggregation weights are dynamically updated based on the model discrepancy. We evaluated our method on two tasks and two types of data, graph generation and image classification (binary and multi-class), with both homogeneous and heterogeneous model architectures. Experimental results demonstrate the effectiveness and outperformance of RepTreeFL in settings where both data and clients are limited. Our code is available at https://github.com/basiralab/RepTreeFL.
FALCON: Feature-Label Constrained Graph Net Collapse for Memory Efficient GNNs
Dec 27, 2023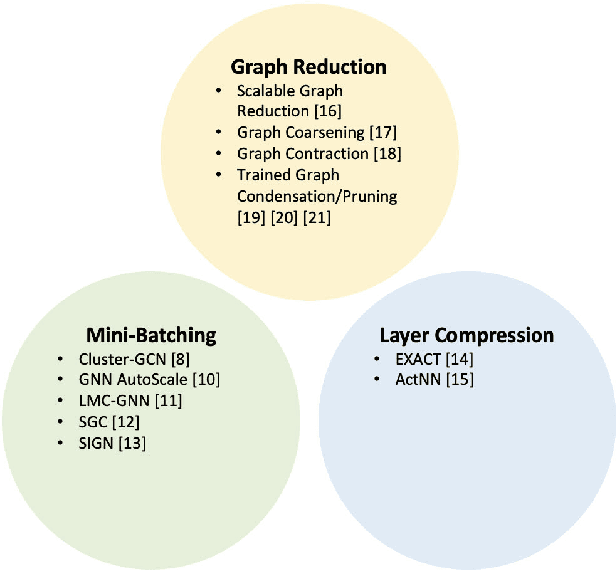
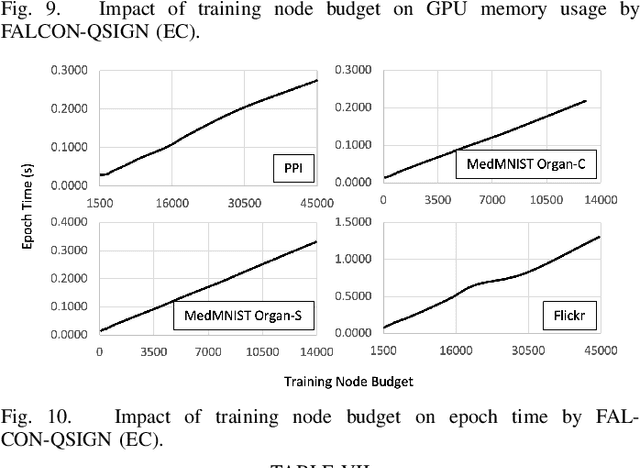
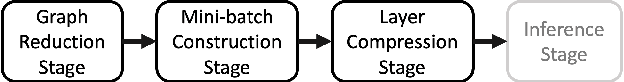
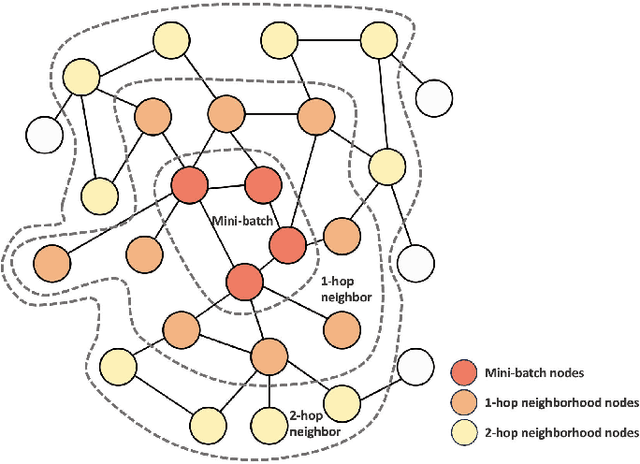
Abstract:Graph Neural Network (GNN) ushered in a new era of machine learning with interconnected datasets. While traditional neural networks can only be trained on independent samples, GNN allows for the inclusion of inter-sample interactions in the training process. This gain, however, incurs additional memory cost, rendering most GNNs unscalable for real-world applications involving vast and complicated networks with tens of millions of nodes (e.g., social circles, web graphs, and brain graphs). This means that storing the graph in the main memory can be difficult, let alone training the GNN model with significantly less GPU memory. While much of the recent literature has focused on either mini-batching GNN methods or quantization, graph reduction methods remain largely scarce. Furthermore, present graph reduction approaches have several drawbacks. First, most graph reduction focuses only on the inference stage (e.g., condensation and distillation) and requires full graph GNN training, which does not reduce training memory footprint. Second, many methods focus solely on the graph's structural aspect, ignoring the initial population feature-label distribution, resulting in a skewed post-reduction label distribution. Here, we propose a Feature-Label COnstrained graph Net collapse, FALCON, to address these limitations. Our three core contributions lie in (i) designing FALCON, a topology-aware graph reduction technique that preserves feature-label distribution; (ii) implementation of FALCON with other memory reduction methods (i.e., mini-batched GNN and quantization) for further memory reduction; (iii) extensive benchmarking and ablation studies against SOTA methods to evaluate FALCON memory reduction. Our extensive results show that FALCON can significantly collapse various public datasets while achieving equal prediction quality across GNN models. Code: https://github.com/basiralab/FALCON
Foundational Models in Medical Imaging: A Comprehensive Survey and Future Vision
Oct 28, 2023



Abstract:Foundation models, large-scale, pre-trained deep-learning models adapted to a wide range of downstream tasks have gained significant interest lately in various deep-learning problems undergoing a paradigm shift with the rise of these models. Trained on large-scale dataset to bridge the gap between different modalities, foundation models facilitate contextual reasoning, generalization, and prompt capabilities at test time. The predictions of these models can be adjusted for new tasks by augmenting the model input with task-specific hints called prompts without requiring extensive labeled data and retraining. Capitalizing on the advances in computer vision, medical imaging has also marked a growing interest in these models. To assist researchers in navigating this direction, this survey intends to provide a comprehensive overview of foundation models in the domain of medical imaging. Specifically, we initiate our exploration by providing an exposition of the fundamental concepts forming the basis of foundation models. Subsequently, we offer a methodical taxonomy of foundation models within the medical domain, proposing a classification system primarily structured around training strategies, while also incorporating additional facets such as application domains, imaging modalities, specific organs of interest, and the algorithms integral to these models. Furthermore, we emphasize the practical use case of some selected approaches and then discuss the opportunities, applications, and future directions of these large-scale pre-trained models, for analyzing medical images. In the same vein, we address the prevailing challenges and research pathways associated with foundational models in medical imaging. These encompass the areas of interpretability, data management, computational requirements, and the nuanced issue of contextual comprehension.
Population Template-Based Brain Graph Augmentation for Improving One-Shot Learning Classification
Dec 14, 2022Abstract:The challenges of collecting medical data on neurological disorder diagnosis problems paved the way for learning methods with scarce number of samples. Due to this reason, one-shot learning still remains one of the most challenging and trending concepts of deep learning as it proposes to simulate the human-like learning approach in classification problems. Previous studies have focused on generating more accurate fingerprints of the population using graph neural networks (GNNs) with connectomic brain graph data. Thereby, generated population fingerprints named connectional brain template (CBTs) enabled detecting discriminative bio-markers of the population on classification tasks. However, the reverse problem of data augmentation from single graph data representing brain connectivity has never been tackled before. In this paper, we propose an augmentation pipeline in order to provide improved metrics on our binary classification problem. Divergently from the previous studies, we examine augmentation from a single population template by utilizing graph-based generative adversarial network (gGAN) architecture for a classification problem. We benchmarked our proposed solution on AD/LMCI dataset consisting of brain connectomes with Alzheimer's Disease (AD) and Late Mild Cognitive Impairment (LMCI). In order to evaluate our model's generalizability, we used cross-validation strategy and randomly sampled the folds multiple times. Our results on classification not only provided better accuracy when augmented data generated from one sample is introduced, but yields more balanced results on other metrics as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge