Weili Lin
Senior Member, IEEE
Predicting Infant Brain Connectivity with Federated Multi-Trajectory GNNs using Scarce Data
Jan 08, 2024



Abstract:The understanding of the convoluted evolution of infant brain networks during the first postnatal year is pivotal for identifying the dynamics of early brain connectivity development. Existing deep learning solutions suffer from three major limitations. First, they cannot generalize to multi-trajectory prediction tasks, where each graph trajectory corresponds to a particular imaging modality or connectivity type (e.g., T1-w MRI). Second, existing models require extensive training datasets to achieve satisfactory performance which are often challenging to obtain. Third, they do not efficiently utilize incomplete time series data. To address these limitations, we introduce FedGmTE-Net++, a federated graph-based multi-trajectory evolution network. Using the power of federation, we aggregate local learnings among diverse hospitals with limited datasets. As a result, we enhance the performance of each hospital's local generative model, while preserving data privacy. The three key innovations of FedGmTE-Net++ are: (i) presenting the first federated learning framework specifically designed for brain multi-trajectory evolution prediction in a data-scarce environment, (ii) incorporating an auxiliary regularizer in the local objective function to exploit all the longitudinal brain connectivity within the evolution trajectory and maximize data utilization, (iii) introducing a two-step imputation process, comprising a preliminary KNN-based precompletion followed by an imputation refinement step that employs regressors to improve similarity scores and refine imputations. Our comprehensive experimental results showed the outperformance of FedGmTE-Net++ in brain multi-trajectory prediction from a single baseline graph in comparison with benchmark methods.
Source-Free Unsupervised Domain Adaptation: A Survey
Jan 06, 2023



Abstract:Unsupervised domain adaptation (UDA) via deep learning has attracted appealing attention for tackling domain-shift problems caused by distribution discrepancy across different domains. Existing UDA approaches highly depend on the accessibility of source domain data, which is usually limited in practical scenarios due to privacy protection, data storage and transmission cost, and computation burden. To tackle this issue, many source-free unsupervised domain adaptation (SFUDA) methods have been proposed recently, which perform knowledge transfer from a pre-trained source model to unlabeled target domain with source data inaccessible. A comprehensive review of these works on SFUDA is of great significance. In this paper, we provide a timely and systematic literature review of existing SFUDA approaches from a technical perspective. Specifically, we categorize current SFUDA studies into two groups, i.e., white-box SFUDA and black-box SFUDA, and further divide them into finer subcategories based on different learning strategies they use. We also investigate the challenges of methods in each subcategory, discuss the advantages/disadvantages of white-box and black-box SFUDA methods, conclude the commonly used benchmark datasets, and summarize the popular techniques for improved generalizability of models learned without using source data. We finally discuss several promising future directions in this field.
Longitudinal Prediction of Postnatal Brain Magnetic Resonance Images via a Metamorphic Generative Adversarial Network
Aug 09, 2022



Abstract:Missing scans are inevitable in longitudinal studies due to either subject dropouts or failed scans. In this paper, we propose a deep learning framework to predict missing scans from acquired scans, catering to longitudinal infant studies. Prediction of infant brain MRI is challenging owing to the rapid contrast and structural changes particularly during the first year of life. We introduce a trustworthy metamorphic generative adversarial network (MGAN) for translating infant brain MRI from one time-point to another. MGAN has three key features: (i) Image translation leveraging spatial and frequency information for detail-preserving mapping; (ii) Quality-guided learning strategy that focuses attention on challenging regions. (iii) Multi-scale hybrid loss function that improves translation of tissue contrast and structural details. Experimental results indicate that MGAN outperforms existing GANs by accurately predicting both contrast and anatomical details.
Learning MRI Artifact Removal With Unpaired Data
Oct 09, 2021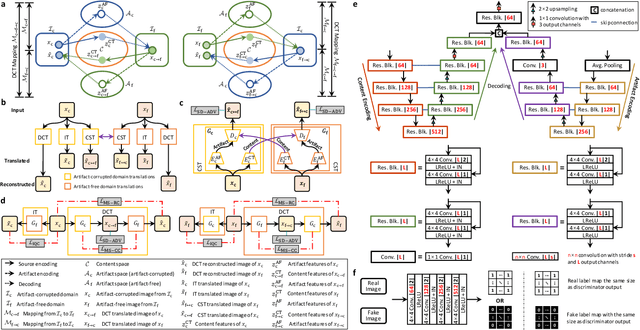
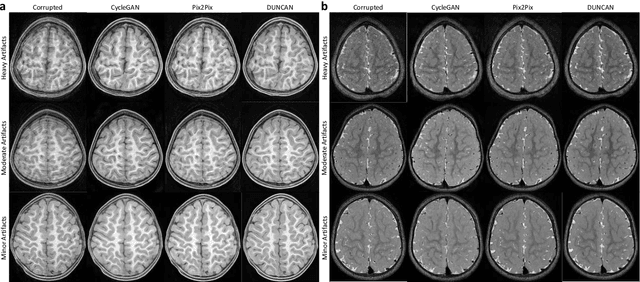
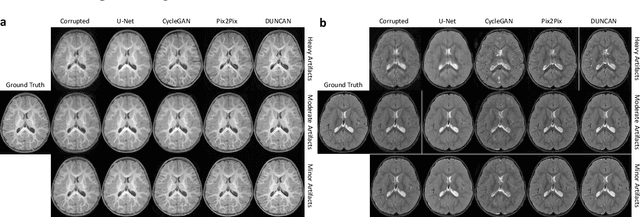
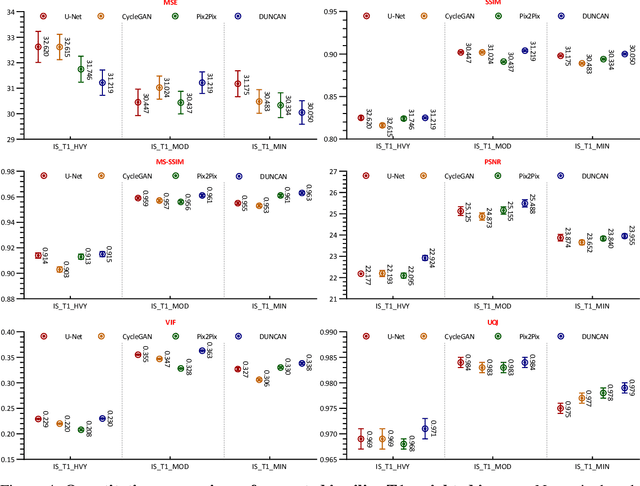
Abstract:Retrospective artifact correction (RAC) improves image quality post acquisition and enhances image usability. Recent machine learning driven techniques for RAC are predominantly based on supervised learning and therefore practical utility can be limited as data with paired artifact-free and artifact-corrupted images are typically insufficient or even non-existent. Here we show that unwanted image artifacts can be disentangled and removed from an image via an RAC neural network learned with unpaired data. This implies that our method does not require matching artifact-corrupted data to be either collected via acquisition or generated via simulation. Experimental results demonstrate that our method is remarkably effective in removing artifacts and retaining anatomical details in images with different contrasts.
A Few-shot Learning Graph Multi-Trajectory Evolution Network for Forecasting Multimodal Baby Connectivity Development from a Baseline Timepoint
Oct 06, 2021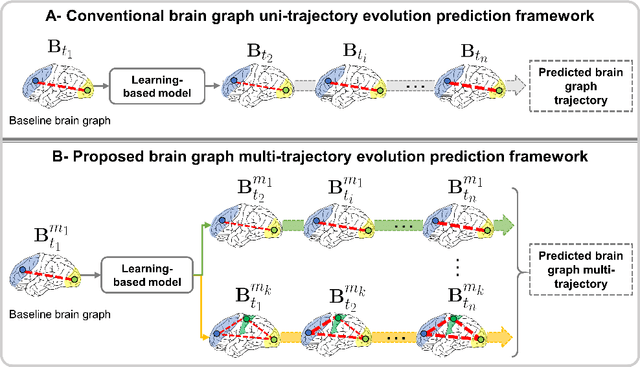
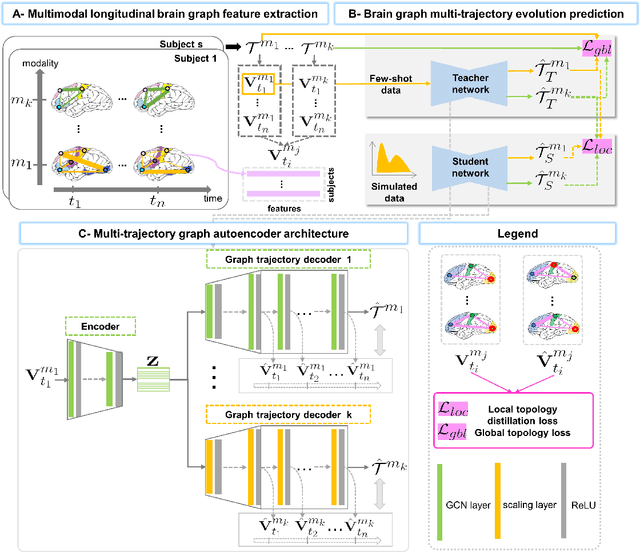
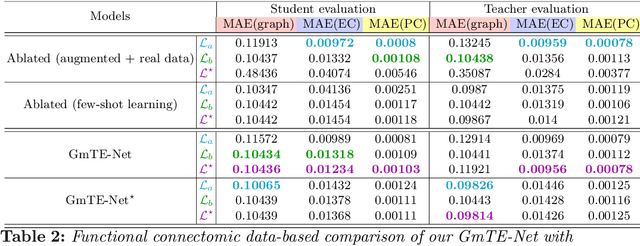
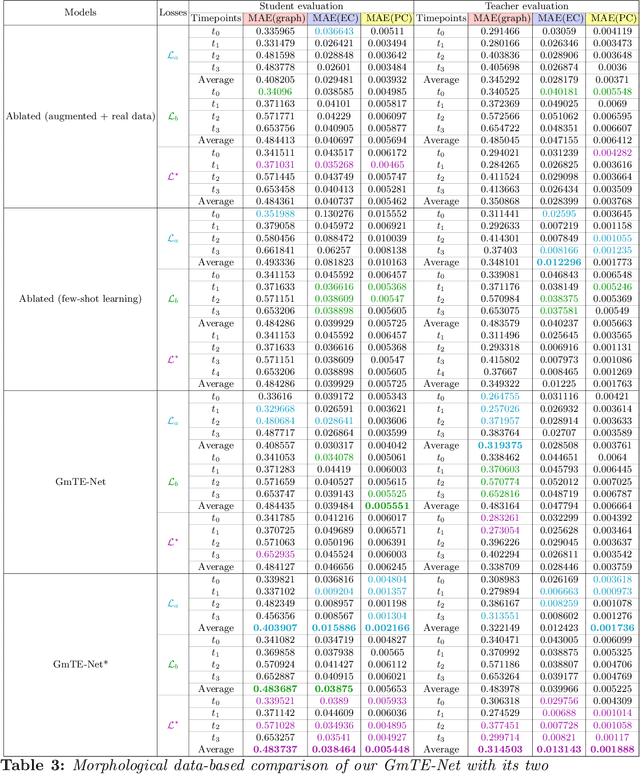
Abstract:Charting the baby connectome evolution trajectory during the first year after birth plays a vital role in understanding dynamic connectivity development of baby brains. Such analysis requires acquisition of longitudinal connectomic datasets. However, both neonatal and postnatal scans are rarely acquired due to various difficulties. A small body of works has focused on predicting baby brain evolution trajectory from a neonatal brain connectome derived from a single modality. Although promising, large training datasets are essential to boost model learning and to generalize to a multi-trajectory prediction from different modalities (i.e., functional and morphological connectomes). Here, we unprecedentedly explore the question: Can we design a few-shot learning-based framework for predicting brain graph trajectories across different modalities? To this aim, we propose a Graph Multi-Trajectory Evolution Network (GmTE-Net), which adopts a teacher-student paradigm where the teacher network learns on pure neonatal brain graphs and the student network learns on simulated brain graphs given a set of different timepoints. To the best of our knowledge, this is the first teacher-student architecture tailored for brain graph multi-trajectory growth prediction that is based on few-shot learning and generalized to graph neural networks (GNNs). To boost the performance of the student network, we introduce a local topology-aware distillation loss that forces the predicted graph topology of the student network to be consistent with the teacher network. Experimental results demonstrate substantial performance gains over benchmark methods. Hence, our GmTE-Net can be leveraged to predict atypical brain connectivity trajectory evolution across various modalities. Our code is available at https: //github.com/basiralab/GmTE-Net.
Co-evolution of Functional Brain Network at Multiple Scales during Early Infancy
Sep 15, 2020



Abstract:The human brains are organized into hierarchically modular networks facilitating efficient and stable information processing and supporting diverse cognitive processes during the course of development. While the remarkable reconfiguration of functional brain network has been firmly established in early life, all these studies investigated the network development from a "single-scale" perspective, which ignore the richness engendered by its hierarchical nature. To fill this gap, this paper leveraged a longitudinal infant resting-state functional magnetic resonance imaging dataset from birth to 2 years of age, and proposed an advanced methodological framework to delineate the multi-scale reconfiguration of functional brain network during early development. Our proposed framework is consist of two parts. The first part developed a novel two-step multi-scale module detection method that could uncover efficient and consistent modular structure for longitudinal dataset from multiple scales in a completely data-driven manner. The second part designed a systematic approach that employed the linear mixed-effect model to four global and nodal module-related metrics to delineate scale-specific age-related changes of network organization. By applying our proposed methodological framework on the collected longitudinal infant dataset, we provided the first evidence that, in the first 2 years of life, the brain functional network is co-evolved at different scales, where each scale displays the unique reconfiguration pattern in terms of modular organization.
Multi-Site Infant Brain Segmentation Algorithms: The iSeg-2019 Challenge
Jul 11, 2020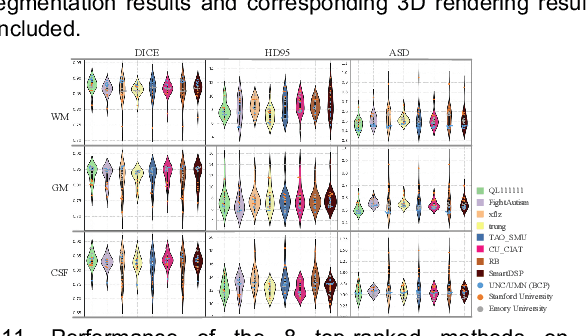
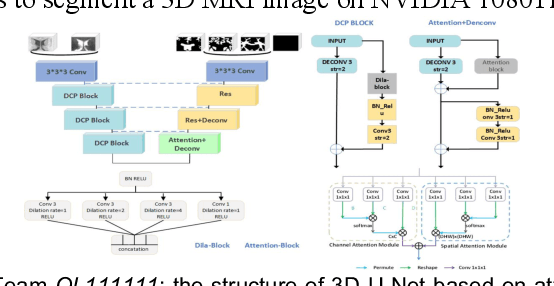
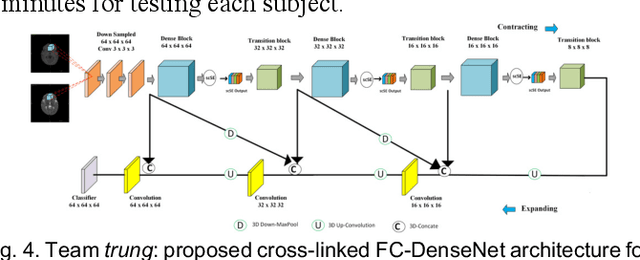
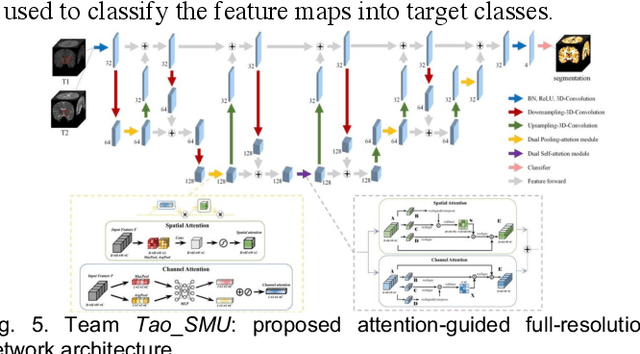
Abstract:To better understand early brain growth patterns in health and disorder, it is critical to accurately segment infant brain magnetic resonance (MR) images into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF). Deep learning-based methods have achieved state-of-the-art performance; however, one of major limitations is that the learning-based methods may suffer from the multi-site issue, that is, the models trained on a dataset from one site may not be applicable to the datasets acquired from other sites with different imaging protocols/scanners. To promote methodological development in the community, iSeg-2019 challenge (http://iseg2019.web.unc.edu) provides a set of 6-month infant subjects from multiple sites with different protocols/scanners for the participating methods. Training/validation subjects are from UNC (MAP) and testing subjects are from UNC/UMN (BCP), Stanford University, and Emory University. By the time of writing, there are 30 automatic segmentation methods participating in iSeg-2019. We review the 8 top-ranked teams by detailing their pipelines/implementations, presenting experimental results and evaluating performance in terms of the whole brain, regions of interest, and gyral landmark curves. We also discuss their limitations and possible future directions for the multi-site issue. We hope that the multi-site dataset in iSeg-2019 and this review article will attract more researchers on the multi-site issue.
Multifold Acceleration of Diffusion MRI via Slice-Interleaved Diffusion Encoding (SIDE)
Feb 25, 2020



Abstract:Diffusion MRI (dMRI) is a unique imaging technique for in vivo characterization of tissue microstructure and white matter pathways. However, its relatively long acquisition time implies greater motion artifacts when imaging, for example, infants and Parkinson's disease patients. To accelerate dMRI acquisition, we propose in this paper (i) a diffusion encoding scheme, called Slice-Interleaved Diffusion Encoding (SIDE), that interleaves each diffusion-weighted (DW) image volume with slices that are encoded with different diffusion gradients, essentially allowing the slice-undersampling of image volume associated with each diffusion gradient to significantly reduce acquisition time, and (ii) a method based on deep learning for effective reconstruction of DW images from the highly slice-undersampled data. Evaluation based on the Human Connectome Project (HCP) dataset indicates that our method can achieve a high acceleration factor of up to 6 with minimal information loss. Evaluation using dMRI data acquired with SIDE acquisition demonstrates that it is possible to accelerate the acquisition by as much as 50 folds when combined with multi-band imaging.
FRNET: Flattened Residual Network for Infant MRI Skull Stripping
Apr 11, 2019



Abstract:Skull stripping for brain MR images is a basic segmentation task. Although many methods have been proposed, most of them focused mainly on the adult MR images. Skull stripping for infant MR images is more challenging due to the small size and dynamic intensity changes of brain tissues during the early ages. In this paper, we propose a novel CNN based framework to robustly extract brain region from infant MR image without any human assistance. Specifically, we propose a simplified but more robust flattened residual network architecture (FRnet). We also introduce a new boundary loss function to highlight ambiguous and low contrast regions between brain and non-brain regions. To make the whole framework more robust to MR images with different imaging quality, we further introduce an artifact simulator for data augmentation. We have trained and tested our proposed framework on a large dataset (N=343), covering newborns to 48-month-olds, and obtained performance better than the state-of-the-art methods in all age groups.
Real-Time Quality Assessment of Pediatric MRI via Semi-Supervised Deep Nonlocal Residual Neural Networks
Apr 07, 2019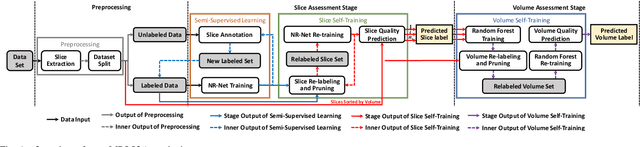
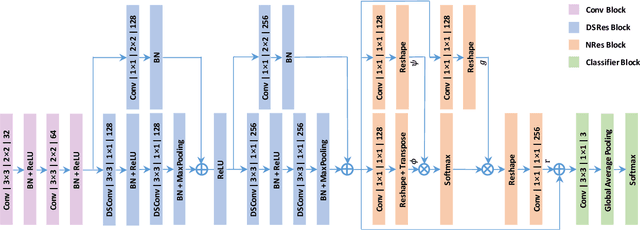
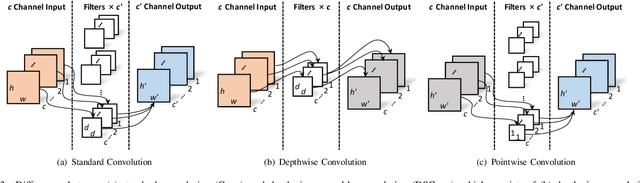
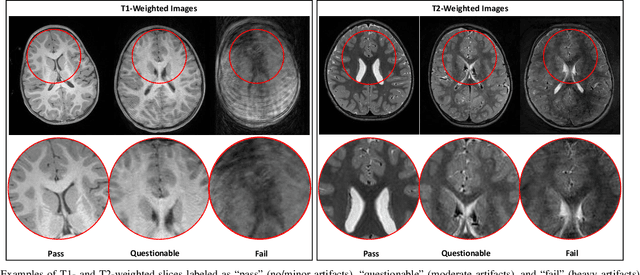
Abstract:In this paper, we introduce an image quality assessment (IQA) method for pediatric T1- and T2-weighted MR images. IQA is first performed slice-wise using a nonlocal residual neural network (NR-Net) and then volume-wise by agglomerating the slice QA results using random forest. Our method requires only a small amount of quality-annotated images for training and is designed to be robust to annotation noise that might occur due to rater errors and the inevitable mix of good and bad slices in an image volume. Using a small set of quality-assessed images, we pre-train NR-Net to annotate each image slice with an initial quality rating (i.e., pass, questionable, fail), which we then refine by semi-supervised learning and iterative self-training. Experimental results demonstrate that our method, trained using only samples of modest size, exhibit great generalizability, capable of real-time (milliseconds per volume) large-scale IQA with near-perfect accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge