Mohamed Ali Mahjoub
SAGE
Jedi: Entropy-based Localization and Removal of Adversarial Patches
Apr 20, 2023



Abstract:Real-world adversarial physical patches were shown to be successful in compromising state-of-the-art models in a variety of computer vision applications. Existing defenses that are based on either input gradient or features analysis have been compromised by recent GAN-based attacks that generate naturalistic patches. In this paper, we propose Jedi, a new defense against adversarial patches that is resilient to realistic patch attacks. Jedi tackles the patch localization problem from an information theory perspective; leverages two new ideas: (1) it improves the identification of potential patch regions using entropy analysis: we show that the entropy of adversarial patches is high, even in naturalistic patches; and (2) it improves the localization of adversarial patches, using an autoencoder that is able to complete patch regions from high entropy kernels. Jedi achieves high-precision adversarial patch localization, which we show is critical to successfully repair the images. Since Jedi relies on an input entropy analysis, it is model-agnostic, and can be applied on pre-trained off-the-shelf models without changes to the training or inference of the protected models. Jedi detects on average 90% of adversarial patches across different benchmarks and recovers up to 94% of successful patch attacks (Compared to 75% and 65% for LGS and Jujutsu, respectively).
Adversarial Attacks in a Multi-view Setting: An Empirical Study of the Adversarial Patches Inter-view Transferability
Oct 10, 2021

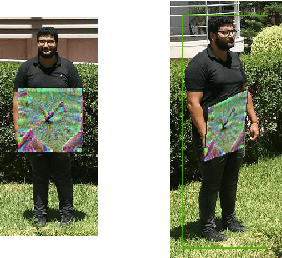

Abstract:While machine learning applications are getting mainstream owing to a demonstrated efficiency in solving complex problems, they suffer from inherent vulnerability to adversarial attacks. Adversarial attacks consist of additive noise to an input which can fool a detector. Recently, successful real-world printable adversarial patches were proven efficient against state-of-the-art neural networks. In the transition from digital noise based attacks to real-world physical attacks, the myriad of factors affecting object detection will also affect adversarial patches. Among these factors, view angle is one of the most influential, yet under-explored. In this paper, we study the effect of view angle on the effectiveness of an adversarial patch. To this aim, we propose the first approach that considers a multi-view context by combining existing adversarial patches with a perspective geometric transformation in order to simulate the effect of view angle changes. Our approach has been evaluated on two datasets: the first dataset which contains most real world constraints of a multi-view context, and the second dataset which empirically isolates the effect of view angle. The experiments show that view angle significantly affects the performance of adversarial patches, where in some cases the patch loses most of its effectiveness. We believe that these results motivate taking into account the effect of view angles in future adversarial attacks, and open up new opportunities for adversarial defenses.
StairwayGraphNet for Inter- and Intra-modality Multi-resolution Brain Graph Alignment and Synthesis
Oct 06, 2021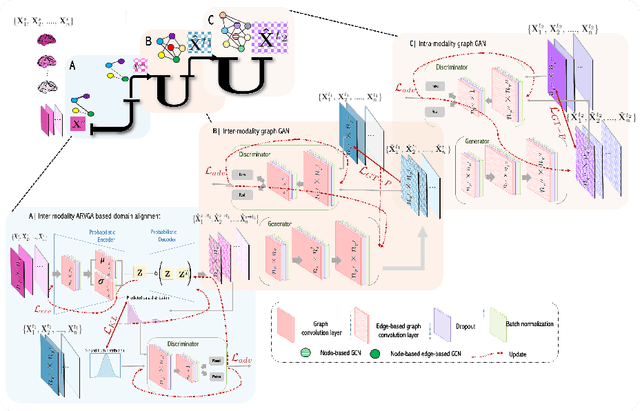
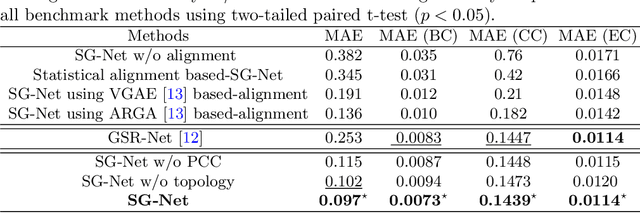

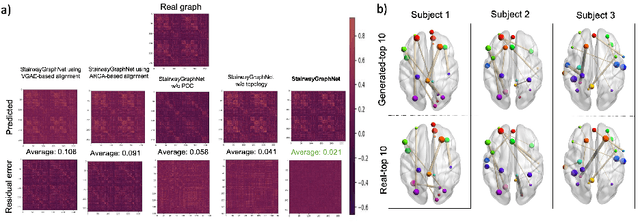
Abstract:Synthesizing multimodality medical data provides complementary knowledge and helps doctors make precise clinical decisions. Although promising, existing multimodal brain graph synthesis frameworks have several limitations. First, they mainly tackle only one problem (intra- or inter-modality), limiting their generalizability to synthesizing inter- and intra-modality simultaneously. Second, while few techniques work on super-resolving low-resolution brain graphs within a single modality (i.e., intra), inter-modality graph super-resolution remains unexplored though this would avoid the need for costly data collection and processing. More importantly, both target and source domains might have different distributions, which causes a domain fracture between them. To fill these gaps, we propose a multi-resolution StairwayGraphNet (SG-Net) framework to jointly infer a target graph modality based on a given modality and super-resolve brain graphs in both inter and intra domains. Our SG-Net is grounded in three main contributions: (i) predicting a target graph from a source one based on a novel graph generative adversarial network in both inter (e.g., morphological-functional) and intra (e.g., functional-functional) domains, (ii) generating high-resolution brain graphs without resorting to the time consuming and expensive MRI processing steps, and (iii) enforcing the source distribution to match that of the ground truth graphs using an inter-modality aligner to relax the loss function to optimize. Moreover, we design a new Ground Truth-Preserving loss function to guide both generators in learning the topological structure of ground truth brain graphs more accurately. Our comprehensive experiments on predicting target brain graphs from source graphs using a multi-resolution stairway showed the outperformance of our method in comparison with its variants and state-of-the-art method.
A Few-shot Learning Graph Multi-Trajectory Evolution Network for Forecasting Multimodal Baby Connectivity Development from a Baseline Timepoint
Oct 06, 2021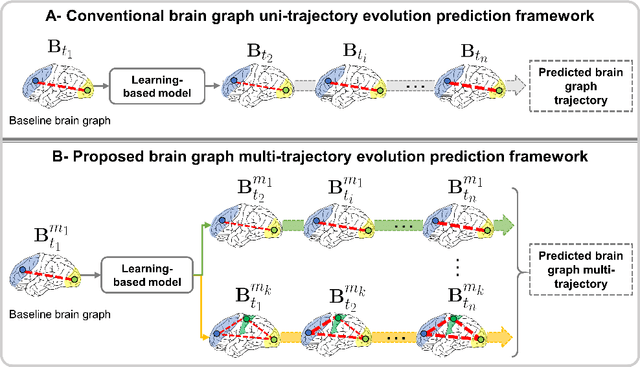
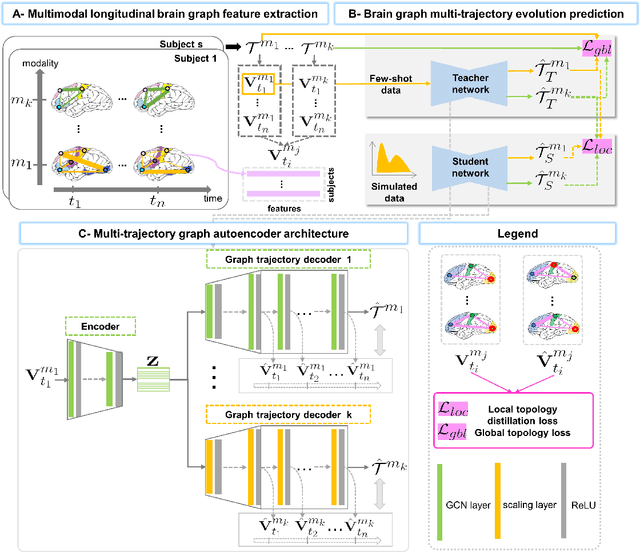
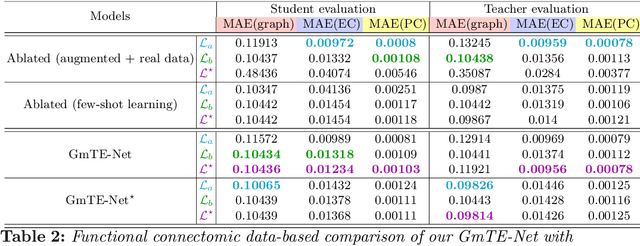
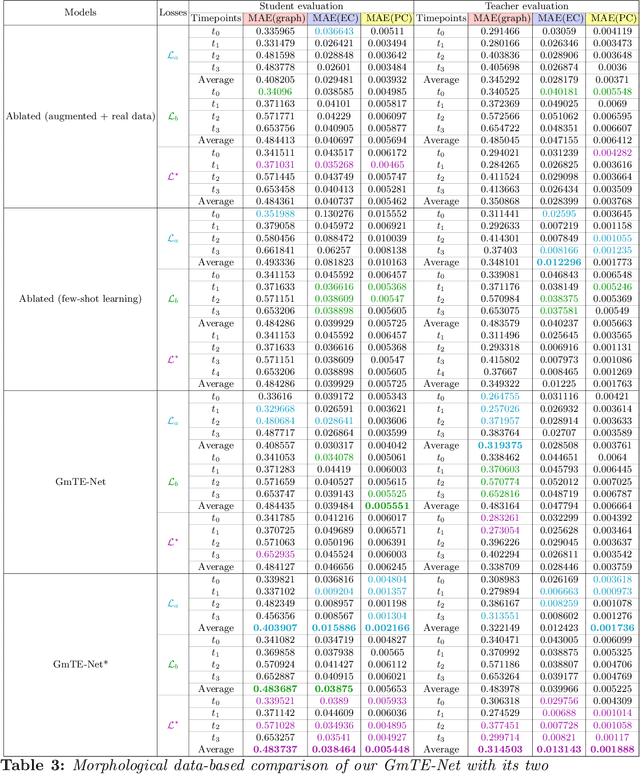
Abstract:Charting the baby connectome evolution trajectory during the first year after birth plays a vital role in understanding dynamic connectivity development of baby brains. Such analysis requires acquisition of longitudinal connectomic datasets. However, both neonatal and postnatal scans are rarely acquired due to various difficulties. A small body of works has focused on predicting baby brain evolution trajectory from a neonatal brain connectome derived from a single modality. Although promising, large training datasets are essential to boost model learning and to generalize to a multi-trajectory prediction from different modalities (i.e., functional and morphological connectomes). Here, we unprecedentedly explore the question: Can we design a few-shot learning-based framework for predicting brain graph trajectories across different modalities? To this aim, we propose a Graph Multi-Trajectory Evolution Network (GmTE-Net), which adopts a teacher-student paradigm where the teacher network learns on pure neonatal brain graphs and the student network learns on simulated brain graphs given a set of different timepoints. To the best of our knowledge, this is the first teacher-student architecture tailored for brain graph multi-trajectory growth prediction that is based on few-shot learning and generalized to graph neural networks (GNNs). To boost the performance of the student network, we introduce a local topology-aware distillation loss that forces the predicted graph topology of the student network to be consistent with the teacher network. Experimental results demonstrate substantial performance gains over benchmark methods. Hence, our GmTE-Net can be leveraged to predict atypical brain connectivity trajectory evolution across various modalities. Our code is available at https: //github.com/basiralab/GmTE-Net.
Non-isomorphic Inter-modality Graph Alignment and Synthesis for Holistic Brain Mapping
Jun 30, 2021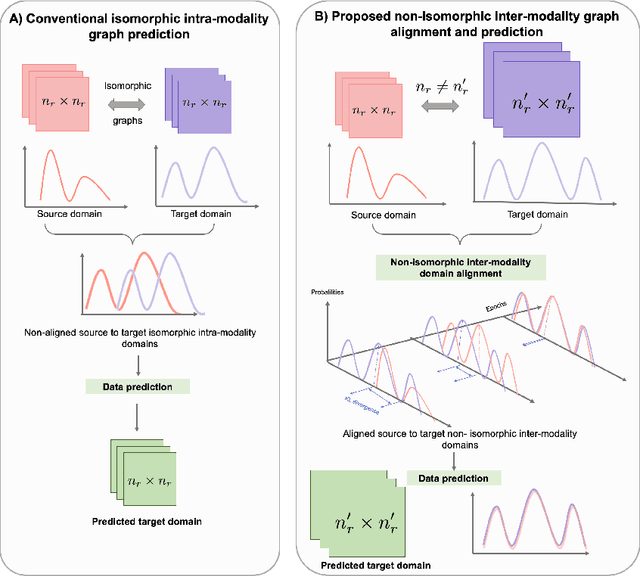
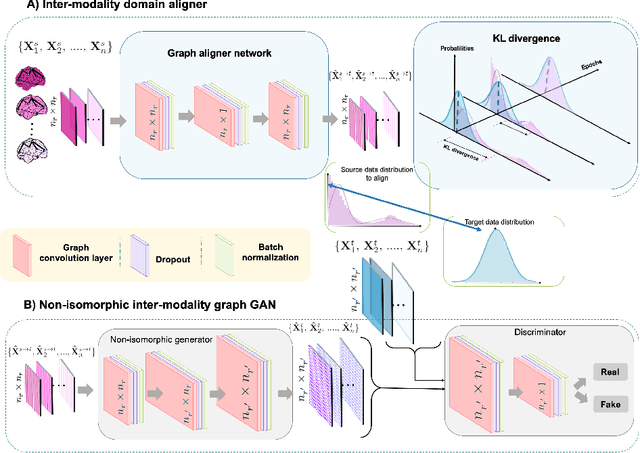
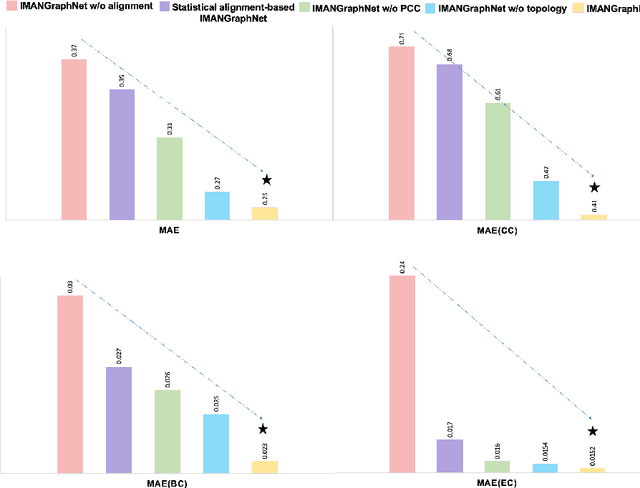
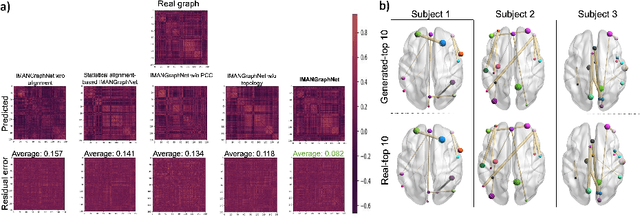
Abstract:Brain graph synthesis marked a new era for predicting a target brain graph from a source one without incurring the high acquisition cost and processing time of neuroimaging data. However, existing multi-modal graph synthesis frameworks have several limitations. First, they mainly focus on generating graphs from the same domain (intra-modality), overlooking the rich multimodal representations of brain connectivity (inter-modality). Second, they can only handle isomorphic graph generation tasks, limiting their generalizability to synthesizing target graphs with a different node size and topological structure from those of the source one. More importantly, both target and source domains might have different distributions, which causes a domain fracture between them (i.e., distribution misalignment). To address such challenges, we propose an inter-modality aligner of non-isomorphic graphs (IMANGraphNet) framework to infer a target graph modality based on a given modality. Our three core contributions lie in (i) predicting a target graph (e.g., functional) from a source graph (e.g., morphological) based on a novel graph generative adversarial network (gGAN); (ii) using non-isomorphic graphs for both source and target domains with a different number of nodes, edges and structure; and (iii) enforcing the predicted target distribution to match that of the ground truth graphs using a graph autoencoder to relax the designed loss oprimization. To handle the unstable behavior of gGAN, we design a new Ground Truth-Preserving (GT-P) loss function to guide the generator in learning the topological structure of ground truth brain graphs. Our comprehensive experiments on predicting functional from morphological graphs demonstrate the outperformance of IMANGraphNet in comparison with its variants. This can be further leveraged for integrative and holistic brain mapping in health and disease.
Graph Neural Networks in Network Neuroscience
Jun 07, 2021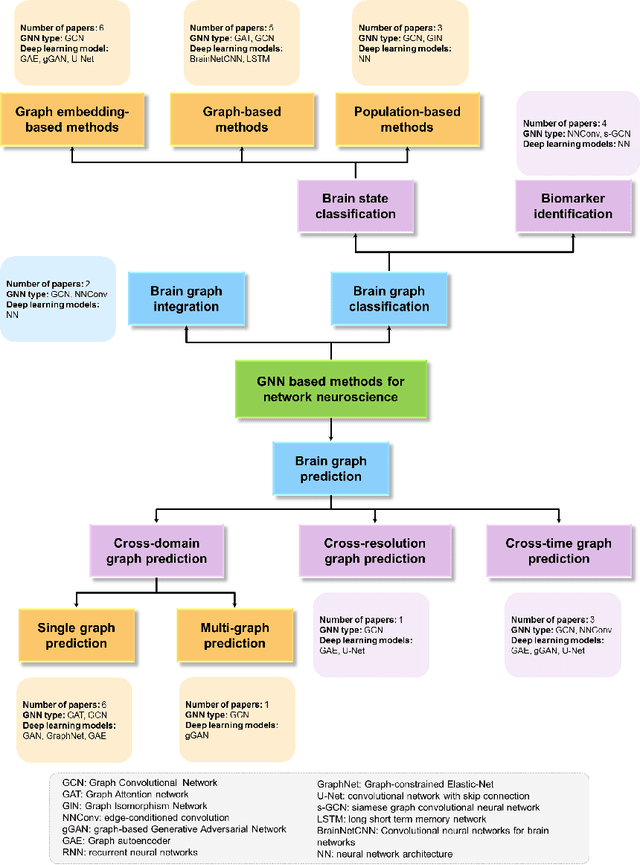

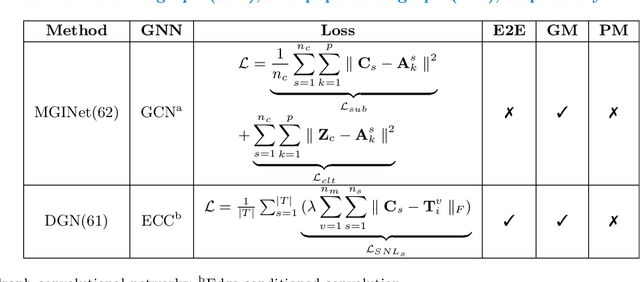
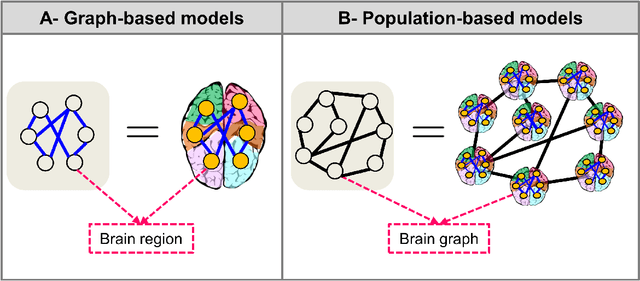
Abstract:Noninvasive medical neuroimaging has yielded many discoveries about the brain connectivity. Several substantial techniques mapping morphological, structural and functional brain connectivities were developed to create a comprehensive road map of neuronal activities in the human brain -namely brain graph. Relying on its non-Euclidean data type, graph neural network (GNN) provides a clever way of learning the deep graph structure and it is rapidly becoming the state-of-the-art leading to enhanced performance in various network neuroscience tasks. Here we review current GNN-based methods, highlighting the ways that they have been used in several applications related to brain graphs such as missing brain graph synthesis and disease classification. We conclude by charting a path toward a better application of GNN models in network neuroscience field for neurological disorder diagnosis and population graph integration. The list of papers cited in our work is available at https://github.com/basiralab/GNNs-in-Network-Neuroscience.
Brain Multigraph Prediction using Topology-Aware Adversarial Graph Neural Network
May 06, 2021

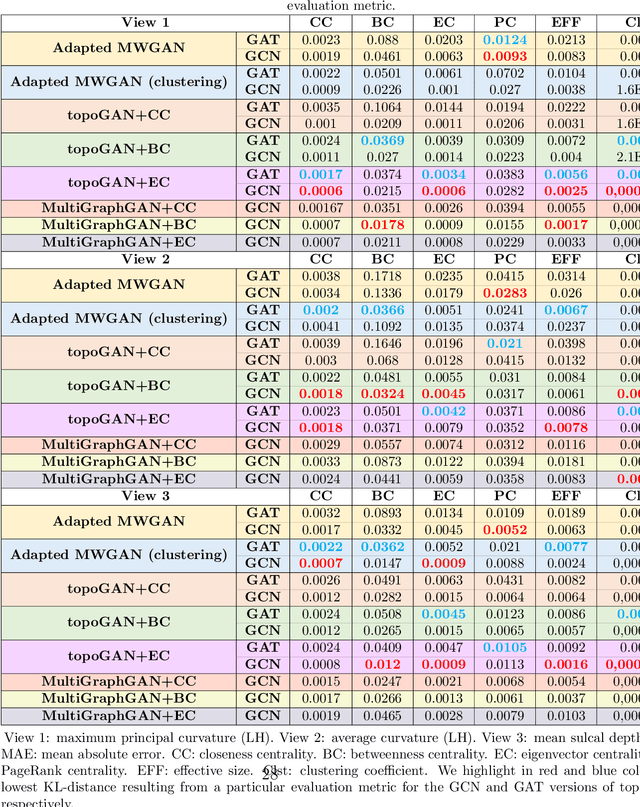
Abstract:Brain graphs (i.e, connectomes) constructed from medical scans such as magnetic resonance imaging (MRI) have become increasingly important tools to characterize the abnormal changes in the human brain. Due to the high acquisition cost and processing time of multimodal MRI, existing deep learning frameworks based on Generative Adversarial Network (GAN) focused on predicting the missing multimodal medical images from a few existing modalities. While brain graphs help better understand how a particular disorder can change the connectional facets of the brain, synthesizing a target brain multigraph (i.e, multiple brain graphs) from a single source brain graph is strikingly lacking. Additionally, existing graph generation works mainly learn one model for each target domain which limits their scalability in jointly predicting multiple target domains. Besides, while they consider the global topological scale of a graph (i.e., graph connectivity structure), they overlook the local topology at the node scale (e.g., how central a node is in the graph). To address these limitations, we introduce topology-aware graph GAN architecture (topoGAN), which jointly predicts multiple brain graphs from a single brain graph while preserving the topological structure of each target graph. Its three key innovations are: (i) designing a novel graph adversarial auto-encoder for predicting multiple brain graphs from a single one, (ii) clustering the encoded source graphs in order to handle the mode collapse issue of GAN and proposing a cluster-specific decoder, (iii) introducing a topological loss to force the prediction of topologically sound target brain graphs. The experimental results using five target domains demonstrated the outperformance of our method in brain multigraph prediction from a single graph in comparison with baseline approaches.
Towards New Multiwavelets: Associated Filters and Algorithms. Part I: Theoretical Framework and Investigation of Biomedical Signals, ECG and Coronavirus Cases
Mar 09, 2021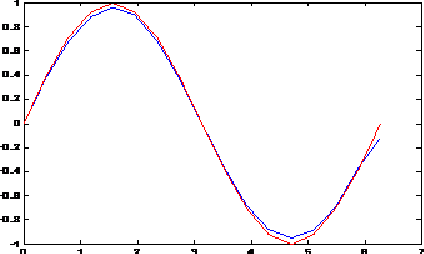

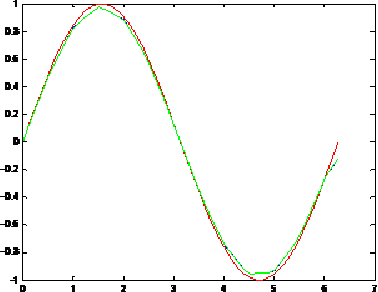
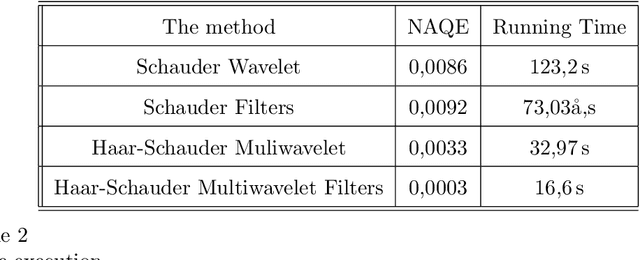
Abstract:Biosignals are nowadays important subjects for scientific researches from both theory and applications especially with the appearance of new pandemics threatening humanity such as the new Coronavirus. One aim in the present work is to prove that Wavelets may be successful machinery to understand such phenomena by applying a step forward extension of wavelets to multiwavelets. We proposed in a first step to improve the multiwavelet notion by constructing more general families using independent components for multi-scaling and multiwavelet mother functions. A special multiwavelet is then introduced, continuous and discrete multiwavelet transforms are associated, as well as new filters and algorithms of decomposition and reconstruction. The constructed multiwavelet framework is applied for some experimentations showing fast algorithms, ECG signal, and a strain of Coronavirus processing.
Topology-Aware Generative Adversarial Network for Joint Prediction of Multiple Brain Graphs from a Single Brain Graph
Sep 23, 2020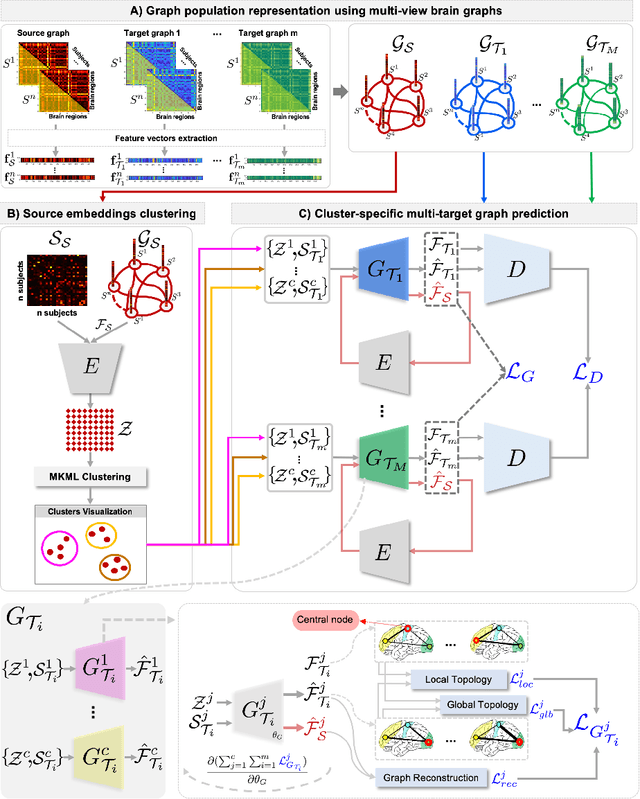

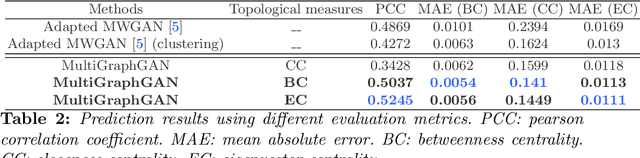
Abstract:Several works based on Generative Adversarial Networks (GAN) have been recently proposed to predict a set of medical images from a single modality (e.g, FLAIR MRI from T1 MRI). However, such frameworks are primarily designed to operate on images, limiting their generalizability to non-Euclidean geometric data such as brain graphs. While a growing number of connectomic studies has demonstrated the promise of including brain graphs for diagnosing neurological disorders, no geometric deep learning work was designed for multiple target brain graphs prediction from a source brain graph. Despite the momentum the field of graph generation has gained in the last two years, existing works have two critical drawbacks. First, the bulk of such works aims to learn one model for each target domain to generate from a source domain. Thus, they have a limited scalability in jointly predicting multiple target domains. Second, they merely consider the global topological scale of a graph (i.e., graph connectivity structure) and overlook the local topology at the node scale of a graph (e.g., how central a node is in the graph). To meet these challenges, we introduce MultiGraphGAN architecture, which not only predicts multiple brain graphs from a single brain graph but also preserves the topological structure of each target graph to predict. Its three core contributions lie in: (i) designing a graph adversarial auto-encoder for jointly predicting brain graphs from a single one, (ii) handling the mode collapse problem of GAN by clustering the encoded source graphs and proposing a cluster-specific decoder, (iii) introducing a topological loss to force the reconstruction of topologically sound target brain graphs. Our MultiGraphGAN significantly outperformed its variants thereby showing its great potential in multi-view brain graph generation from a single graph.
Supervised Multi-topology Network Cross-diffusion for Population-driven Brain Network Atlas Estimation
Sep 23, 2020
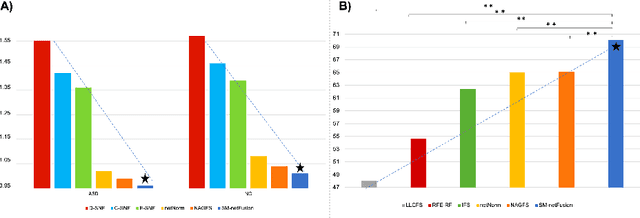

Abstract:Estimating a representative and discriminative brain network atlas (BNA) is a nascent research field in mapping a population of brain networks in health and disease. Although limited, existing BNA estimation methods have several limitations. First, they primarily rely on a similarity network diffusion and fusion technique, which only considers node degree as a topological measure in the cross-network diffusion process, thereby overlooking rich topological measures of the brain network (e.g., centrality). Second, both diffusion and fusion techniques are implemented in fully unsupervised manner, which might decrease the discriminative power of the estimated BNAs. To fill these gaps, we propose a supervised multi-topology network cross-diffusion (SM-netFusion) framework for estimating a BNA satisfying : (i) well-representativeness (captures shared traits across subjects), (ii) well-centeredness (optimally close to all subjects), and (iii) high discriminativeness (can easily and efficiently identify discriminative brain connections that distinguish between two populations). For a specific class, given the cluster labels of the training data, we learn a weighted combination of the topological diffusion kernels derived from degree, closeness and eigenvector centrality measures in a supervised manner. Specifically, we learn the cross-diffusion process by normalizing the training brain networks using the learned diffusion kernels. Our SM-netFusion produces the most centered and representative template in comparison with its variants and state-of-the-art methods and further boosted the classification of autistic subjects by 5-15%. SM-netFusion presents the first work for supervised network cross-diffusion based on graph topological measures, which can be further leveraged to design an efficient graph feature selection method for training predictive learners in network neuroscience.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge