Islem Mhiri
StairwayGraphNet for Inter- and Intra-modality Multi-resolution Brain Graph Alignment and Synthesis
Oct 06, 2021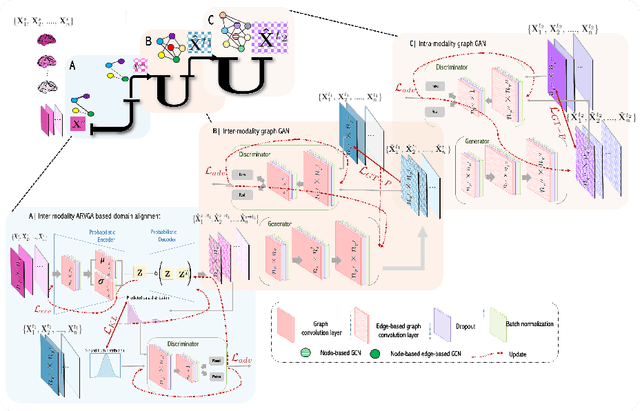
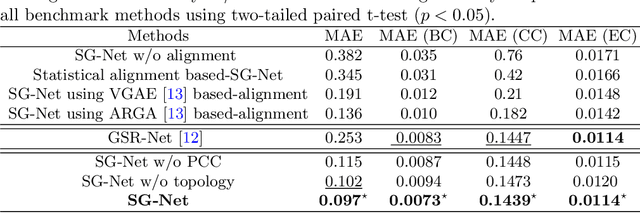

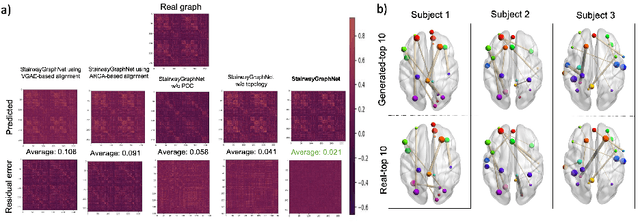
Abstract:Synthesizing multimodality medical data provides complementary knowledge and helps doctors make precise clinical decisions. Although promising, existing multimodal brain graph synthesis frameworks have several limitations. First, they mainly tackle only one problem (intra- or inter-modality), limiting their generalizability to synthesizing inter- and intra-modality simultaneously. Second, while few techniques work on super-resolving low-resolution brain graphs within a single modality (i.e., intra), inter-modality graph super-resolution remains unexplored though this would avoid the need for costly data collection and processing. More importantly, both target and source domains might have different distributions, which causes a domain fracture between them. To fill these gaps, we propose a multi-resolution StairwayGraphNet (SG-Net) framework to jointly infer a target graph modality based on a given modality and super-resolve brain graphs in both inter and intra domains. Our SG-Net is grounded in three main contributions: (i) predicting a target graph from a source one based on a novel graph generative adversarial network in both inter (e.g., morphological-functional) and intra (e.g., functional-functional) domains, (ii) generating high-resolution brain graphs without resorting to the time consuming and expensive MRI processing steps, and (iii) enforcing the source distribution to match that of the ground truth graphs using an inter-modality aligner to relax the loss function to optimize. Moreover, we design a new Ground Truth-Preserving loss function to guide both generators in learning the topological structure of ground truth brain graphs more accurately. Our comprehensive experiments on predicting target brain graphs from source graphs using a multi-resolution stairway showed the outperformance of our method in comparison with its variants and state-of-the-art method.
Non-isomorphic Inter-modality Graph Alignment and Synthesis for Holistic Brain Mapping
Jun 30, 2021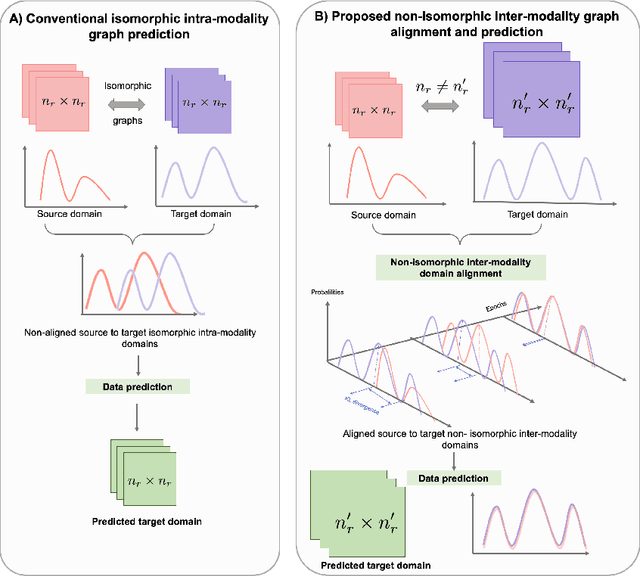
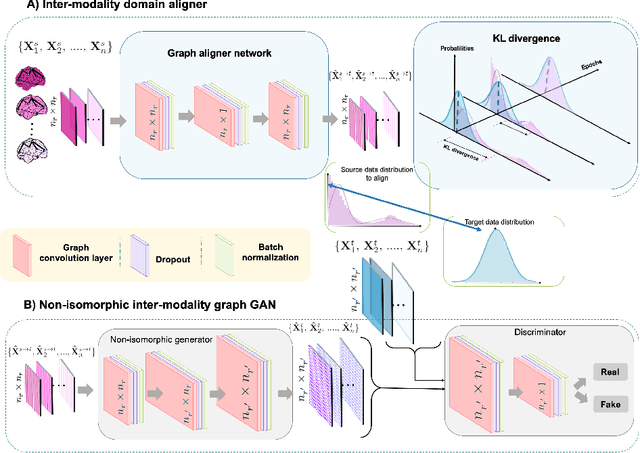
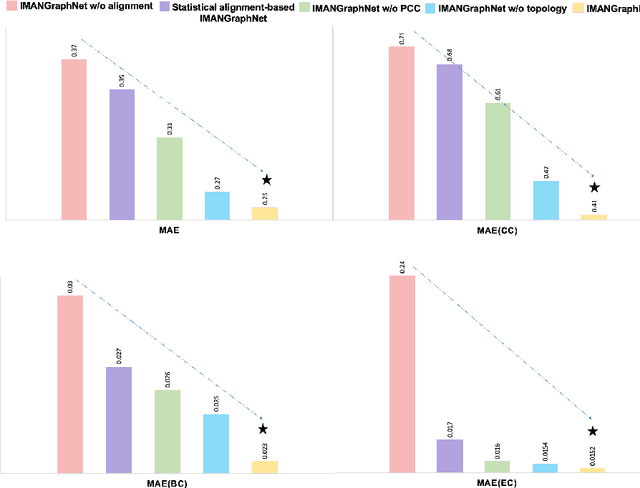
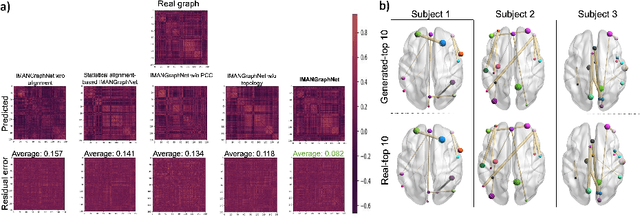
Abstract:Brain graph synthesis marked a new era for predicting a target brain graph from a source one without incurring the high acquisition cost and processing time of neuroimaging data. However, existing multi-modal graph synthesis frameworks have several limitations. First, they mainly focus on generating graphs from the same domain (intra-modality), overlooking the rich multimodal representations of brain connectivity (inter-modality). Second, they can only handle isomorphic graph generation tasks, limiting their generalizability to synthesizing target graphs with a different node size and topological structure from those of the source one. More importantly, both target and source domains might have different distributions, which causes a domain fracture between them (i.e., distribution misalignment). To address such challenges, we propose an inter-modality aligner of non-isomorphic graphs (IMANGraphNet) framework to infer a target graph modality based on a given modality. Our three core contributions lie in (i) predicting a target graph (e.g., functional) from a source graph (e.g., morphological) based on a novel graph generative adversarial network (gGAN); (ii) using non-isomorphic graphs for both source and target domains with a different number of nodes, edges and structure; and (iii) enforcing the predicted target distribution to match that of the ground truth graphs using a graph autoencoder to relax the designed loss oprimization. To handle the unstable behavior of gGAN, we design a new Ground Truth-Preserving (GT-P) loss function to guide the generator in learning the topological structure of ground truth brain graphs. Our comprehensive experiments on predicting functional from morphological graphs demonstrate the outperformance of IMANGraphNet in comparison with its variants. This can be further leveraged for integrative and holistic brain mapping in health and disease.
Supervised Multi-topology Network Cross-diffusion for Population-driven Brain Network Atlas Estimation
Sep 23, 2020
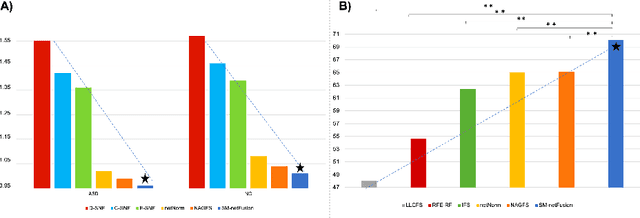

Abstract:Estimating a representative and discriminative brain network atlas (BNA) is a nascent research field in mapping a population of brain networks in health and disease. Although limited, existing BNA estimation methods have several limitations. First, they primarily rely on a similarity network diffusion and fusion technique, which only considers node degree as a topological measure in the cross-network diffusion process, thereby overlooking rich topological measures of the brain network (e.g., centrality). Second, both diffusion and fusion techniques are implemented in fully unsupervised manner, which might decrease the discriminative power of the estimated BNAs. To fill these gaps, we propose a supervised multi-topology network cross-diffusion (SM-netFusion) framework for estimating a BNA satisfying : (i) well-representativeness (captures shared traits across subjects), (ii) well-centeredness (optimally close to all subjects), and (iii) high discriminativeness (can easily and efficiently identify discriminative brain connections that distinguish between two populations). For a specific class, given the cluster labels of the training data, we learn a weighted combination of the topological diffusion kernels derived from degree, closeness and eigenvector centrality measures in a supervised manner. Specifically, we learn the cross-diffusion process by normalizing the training brain networks using the learned diffusion kernels. Our SM-netFusion produces the most centered and representative template in comparison with its variants and state-of-the-art methods and further boosted the classification of autistic subjects by 5-15%. SM-netFusion presents the first work for supervised network cross-diffusion based on graph topological measures, which can be further leveraged to design an efficient graph feature selection method for training predictive learners in network neuroscience.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge