Pew-Thian Yap
Unified Multi-Site Multi-Sequence Brain MRI Harmonization Enriched by Biomedical Semantic Style
Jan 13, 2026Abstract:Aggregating multi-site brain MRI data can enhance deep learning model training, but also introduces non-biological heterogeneity caused by site-specific variations (e.g., differences in scanner vendors, acquisition parameters, and imaging protocols) that can undermine generalizability. Recent retrospective MRI harmonization seeks to reduce such site effects by standardizing image style (e.g., intensity, contrast, noise patterns) while preserving anatomical content. However, existing methods often rely on limited paired traveling-subject data or fail to effectively disentangle style from anatomy. Furthermore, most current approaches address only single-sequence harmonization, restricting their use in real-world settings where multi-sequence MRI is routinely acquired. To this end, we introduce MMH, a unified framework for multi-site multi-sequence brain MRI harmonization that leverages biomedical semantic priors for sequence-aware style alignment. MMH operates in two stages: (1) a diffusion-based global harmonizer that maps MR images to a sequence-specific unified domain using style-agnostic gradient conditioning, and (2) a target-specific fine-tuner that adapts globally aligned images to desired target domains. A tri-planar attention BiomedCLIP encoder aggregates multi-view embeddings to characterize volumetric style information, allowing explicit disentanglement of image styles from anatomy without requiring paired data. Evaluations on 4,163 T1- and T2-weighted MRIs demonstrate MMH's superior harmonization over state-of-the-art methods in image feature clustering, voxel-level comparison, tissue segmentation, and downstream age and site classification.
SinoSynth: A Physics-based Domain Randomization Approach for Generalizable CBCT Image Enhancement
Sep 27, 2024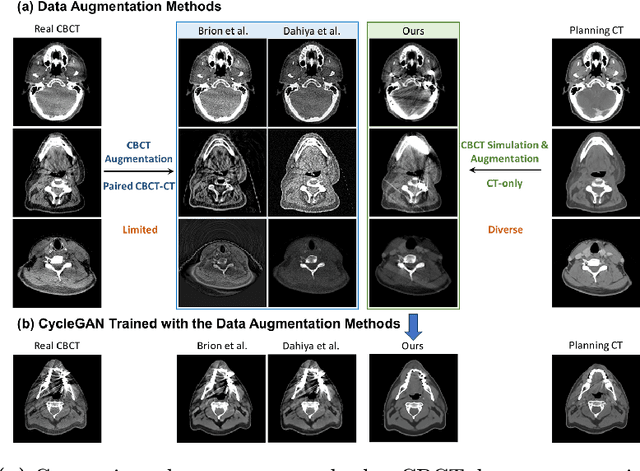
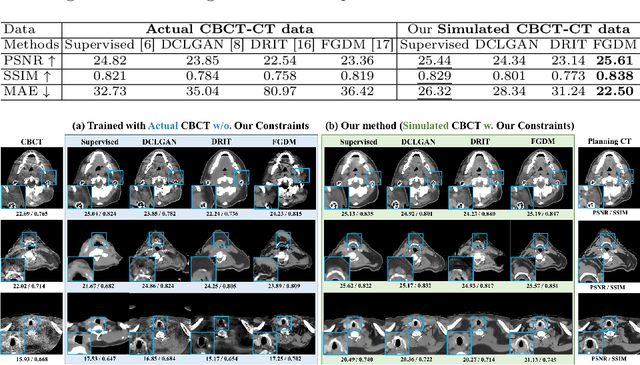
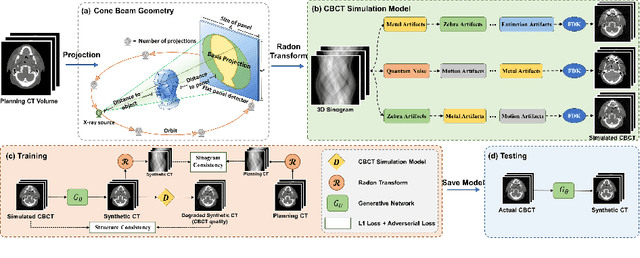
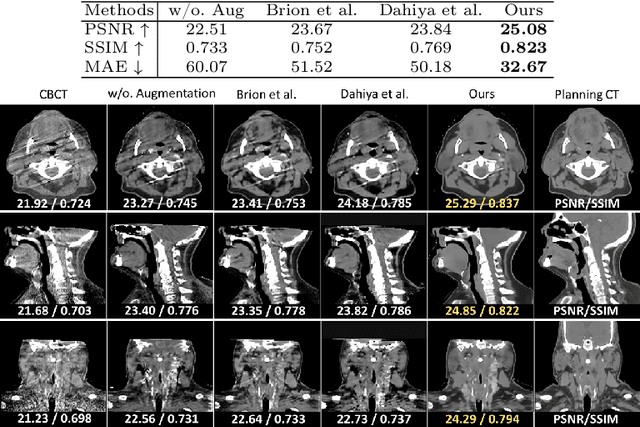
Abstract:Cone Beam Computed Tomography (CBCT) finds diverse applications in medicine. Ensuring high image quality in CBCT scans is essential for accurate diagnosis and treatment delivery. Yet, the susceptibility of CBCT images to noise and artifacts undermines both their usefulness and reliability. Existing methods typically address CBCT artifacts through image-to-image translation approaches. These methods, however, are limited by the artifact types present in the training data, which may not cover the complete spectrum of CBCT degradations stemming from variations in imaging protocols. Gathering additional data to encompass all possible scenarios can often pose a challenge. To address this, we present SinoSynth, a physics-based degradation model that simulates various CBCT-specific artifacts to generate a diverse set of synthetic CBCT images from high-quality CT images without requiring pre-aligned data. Through extensive experiments, we demonstrate that several different generative networks trained on our synthesized data achieve remarkable results on heterogeneous multi-institutional datasets, outperforming even the same networks trained on actual data. We further show that our degradation model conveniently provides an avenue to enforce anatomical constraints in conditional generative models, yielding high-quality and structure-preserving synthetic CT images.
Unpaired Volumetric Harmonization of Brain MRI with Conditional Latent Diffusion
Aug 18, 2024

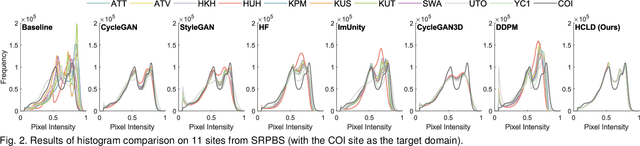

Abstract:Multi-site structural MRI is increasingly used in neuroimaging studies to diversify subject cohorts. However, combining MR images acquired from various sites/centers may introduce site-related non-biological variations. Retrospective image harmonization helps address this issue, but current methods usually perform harmonization on pre-extracted hand-crafted radiomic features, limiting downstream applicability. Several image-level approaches focus on 2D slices, disregarding inherent volumetric information, leading to suboptimal outcomes. To this end, we propose a novel 3D MRI Harmonization framework through Conditional Latent Diffusion (HCLD) by explicitly considering image style and brain anatomy. It comprises a generalizable 3D autoencoder that encodes and decodes MRIs through a 4D latent space, and a conditional latent diffusion model that learns the latent distribution and generates harmonized MRIs with anatomical information from source MRIs while conditioned on target image style. This enables efficient volume-level MRI harmonization through latent style translation, without requiring paired images from target and source domains during training. The HCLD is trained and evaluated on 4,158 T1-weighted brain MRIs from three datasets in three tasks, assessing its ability to remove site-related variations while retaining essential biological features. Qualitative and quantitative experiments suggest the effectiveness of HCLD over several state-of-the-arts
Disentangled Latent Energy-Based Style Translation: An Image-Level Structural MRI Harmonization Framework
Feb 10, 2024



Abstract:Brain magnetic resonance imaging (MRI) has been extensively employed across clinical and research fields, but often exhibits sensitivity to site effects arising from nonbiological variations such as differences in field strength and scanner vendors. Numerous retrospective MRI harmonization techniques have demonstrated encouraging outcomes in reducing the site effects at image level. However, existing methods generally suffer from high computational requirements and limited generalizability, restricting their applicability to unseen MRIs. In this paper, we design a novel disentangled latent energy-based style translation (DLEST) framework for unpaired image-level MRI harmonization, consisting of (1) site-invariant image generation (SIG), (2) site-specific style translation (SST), and (3) site-specific MRI synthesis (SMS). Specifically, the SIG employs a latent autoencoder to encode MRIs into a low-dimensional latent space and reconstruct MRIs from latent codes. The SST utilizes an energy-based model to comprehend the global latent distribution of a target domain and translate source latent codes toward the target domain, while SMS enables MRI synthesis with a target-specific style. By disentangling image generation and style translation in latent space, the DLEST can achieve efficient style translation. Our model was trained on T1-weighted MRIs from a public dataset (with 3,984 subjects across 58 acquisition sites/settings) and validated on an independent dataset (with 9 traveling subjects scanned in 11 sites/settings) in 4 tasks: (1) histogram and clustering comparison, (2) site classification, (3) brain tissue segmentation, and (4) site-specific MRI synthesis. Qualitative and quantitative results demonstrate the superiority of our method over several state-of-the-arts.
Towards Architecture-Insensitive Untrained Network Priors for Accelerated MRI Reconstruction
Dec 15, 2023



Abstract:Untrained neural networks pioneered by Deep Image Prior (DIP) have recently enabled MRI reconstruction without requiring fully-sampled measurements for training. Their success is widely attributed to the implicit regularization induced by suitable network architectures. However, the lack of understanding of such architectural priors results in superfluous design choices and sub-optimal outcomes. This work aims to simplify the architectural design decisions for DIP-MRI to facilitate its practical deployment. We observe that certain architectural components are more prone to causing overfitting regardless of the number of parameters, incurring severe reconstruction artifacts by hindering accurate extrapolation on the un-acquired measurements. We interpret this phenomenon from a frequency perspective and find that the architectural characteristics favoring low frequencies, i.e., deep and narrow with unlearnt upsampling, can lead to enhanced generalization and hence better reconstruction. Building on this insight, we propose two architecture-agnostic remedies: one to constrain the frequency range of the white-noise input and the other to penalize the Lipschitz constants of the network. We demonstrate that even with just one extra line of code on the input, the performance gap between the ill-designed models and the high-performing ones can be closed. These results signify that for the first time, architectural biases on untrained MRI reconstruction can be mitigated without architectural modifications.
Reconstruction of Cortical Surfaces with Spherical Topology from Infant Brain MRI via Recurrent Deformation Learning
Dec 10, 2023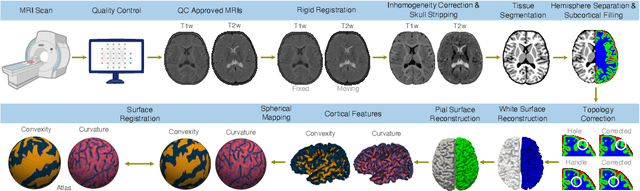
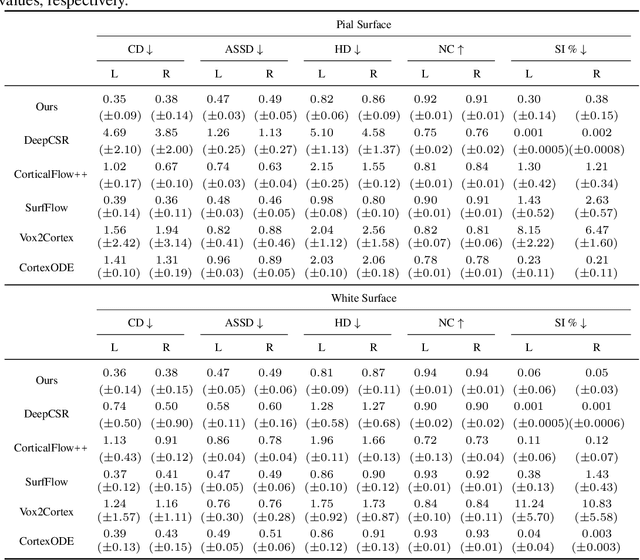
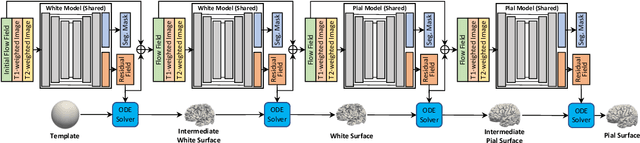
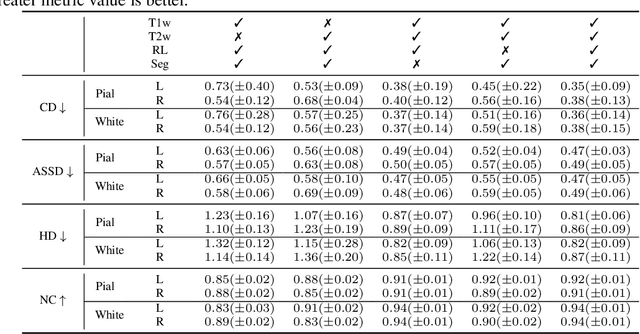
Abstract:Cortical surface reconstruction (CSR) from MRI is key to investigating brain structure and function. While recent deep learning approaches have significantly improved the speed of CSR, a substantial amount of runtime is still needed to map the cortex to a topologically-correct spherical manifold to facilitate downstream geometric analyses. Moreover, this mapping is possible only if the topology of the surface mesh is homotopic to a sphere. Here, we present a method for simultaneous CSR and spherical mapping efficiently within seconds. Our approach seamlessly connects two sub-networks for white and pial surface generation. Residual diffeomorphic deformations are learned iteratively to gradually warp a spherical template mesh to the white and pial surfaces while preserving mesh topology and uniformity. The one-to-one vertex correspondence between the template sphere and the cortical surfaces allows easy and direct mapping of geometric features like convexity and curvature to the sphere for visualization and downstream processing. We demonstrate the efficacy of our approach on infant brain MRI, which poses significant challenges to CSR due to tissue contrast changes associated with rapid brain development during the first postnatal year. Performance evaluation based on a dataset of infants from 0 to 12 months demonstrates that our method substantially enhances mesh regularity and reduces geometric errors, outperforming state-of-the-art deep learning approaches, all while maintaining high computational efficiency.
Source-Free Unsupervised Domain Adaptation: A Survey
Jan 06, 2023



Abstract:Unsupervised domain adaptation (UDA) via deep learning has attracted appealing attention for tackling domain-shift problems caused by distribution discrepancy across different domains. Existing UDA approaches highly depend on the accessibility of source domain data, which is usually limited in practical scenarios due to privacy protection, data storage and transmission cost, and computation burden. To tackle this issue, many source-free unsupervised domain adaptation (SFUDA) methods have been proposed recently, which perform knowledge transfer from a pre-trained source model to unlabeled target domain with source data inaccessible. A comprehensive review of these works on SFUDA is of great significance. In this paper, we provide a timely and systematic literature review of existing SFUDA approaches from a technical perspective. Specifically, we categorize current SFUDA studies into two groups, i.e., white-box SFUDA and black-box SFUDA, and further divide them into finer subcategories based on different learning strategies they use. We also investigate the challenges of methods in each subcategory, discuss the advantages/disadvantages of white-box and black-box SFUDA methods, conclude the commonly used benchmark datasets, and summarize the popular techniques for improved generalizability of models learned without using source data. We finally discuss several promising future directions in this field.
Brain Tissue Segmentation Across the Human Lifespan via Supervised Contrastive Learning
Jan 03, 2023
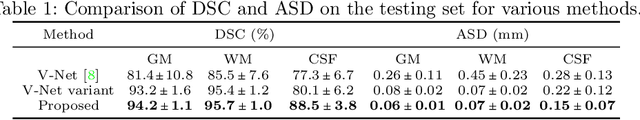
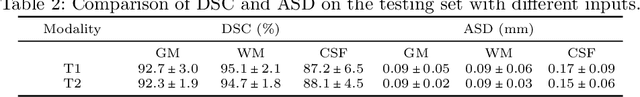
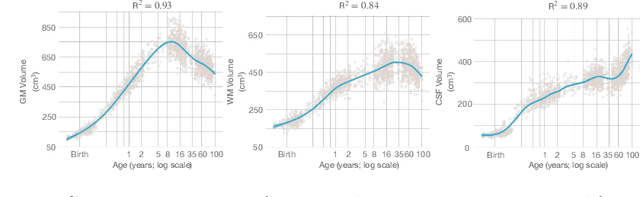
Abstract:Automatic segmentation of brain MR images into white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF) is critical for tissue volumetric analysis and cortical surface reconstruction. Due to dramatic structural and appearance changes associated with developmental and aging processes, existing brain tissue segmentation methods are only viable for specific age groups. Consequently, methods developed for one age group may fail for another. In this paper, we make the first attempt to segment brain tissues across the entire human lifespan (0-100 years of age) using a unified deep learning model. To overcome the challenges related to structural variability underpinned by biological processes, intensity inhomogeneity, motion artifacts, scanner-induced differences, and acquisition protocols, we propose to use contrastive learning to improve the quality of feature representations in a latent space for effective lifespan tissue segmentation. We compared our approach with commonly used segmentation methods on a large-scale dataset of 2,464 MR images. Experimental results show that our model accurately segments brain tissues across the lifespan and outperforms existing methods.
Longitudinal Prediction of Postnatal Brain Magnetic Resonance Images via a Metamorphic Generative Adversarial Network
Aug 09, 2022



Abstract:Missing scans are inevitable in longitudinal studies due to either subject dropouts or failed scans. In this paper, we propose a deep learning framework to predict missing scans from acquired scans, catering to longitudinal infant studies. Prediction of infant brain MRI is challenging owing to the rapid contrast and structural changes particularly during the first year of life. We introduce a trustworthy metamorphic generative adversarial network (MGAN) for translating infant brain MRI from one time-point to another. MGAN has three key features: (i) Image translation leveraging spatial and frequency information for detail-preserving mapping; (ii) Quality-guided learning strategy that focuses attention on challenging regions. (iii) Multi-scale hybrid loss function that improves translation of tissue contrast and structural details. Experimental results indicate that MGAN outperforms existing GANs by accurately predicting both contrast and anatomical details.
Localizing the Recurrent Laryngeal Nerve via Ultrasound with a Bayesian Shape Framework
Jun 30, 2022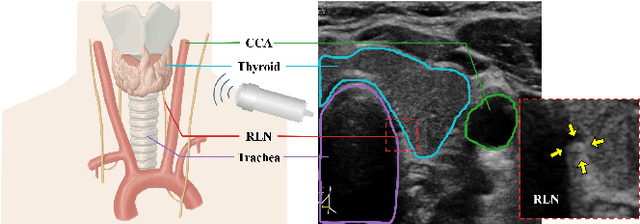
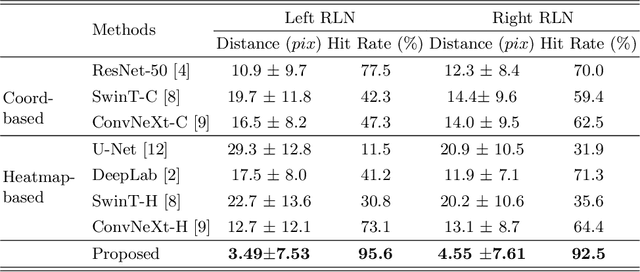
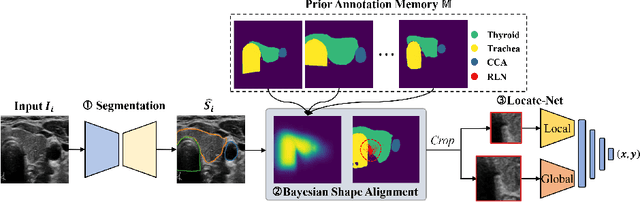
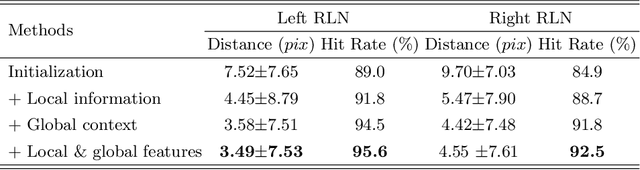
Abstract:Tumor infiltration of the recurrent laryngeal nerve (RLN) is a contraindication for robotic thyroidectomy and can be difficult to detect via standard laryngoscopy. Ultrasound (US) is a viable alternative for RLN detection due to its safety and ability to provide real-time feedback. However, the tininess of the RLN, with a diameter typically less than 3mm, poses significant challenges to the accurate localization of the RLN. In this work, we propose a knowledge-driven framework for RLN localization, mimicking the standard approach surgeons take to identify the RLN according to its surrounding organs. We construct a prior anatomical model based on the inherent relative spatial relationships between organs. Through Bayesian shape alignment (BSA), we obtain the candidate coordinates of the center of a region of interest (ROI) that encloses the RLN. The ROI allows a decreased field of view for determining the refined centroid of the RLN using a dual-path identification network, based on multi-scale semantic information. Experimental results indicate that the proposed method achieves superior hit rates and substantially smaller distance errors compared with state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge