Yilin Liu
Learning to Solve PDEs on Neural Shape Representations
Dec 24, 2025



Abstract:Solving partial differential equations (PDEs) on shapes underpins many shape analysis and engineering tasks; yet, prevailing PDE solvers operate on polygonal/triangle meshes while modern 3D assets increasingly live as neural representations. This mismatch leaves no suitable method to solve surface PDEs directly within the neural domain, forcing explicit mesh extraction or per-instance residual training, preventing end-to-end workflows. We present a novel, mesh-free formulation that learns a local update operator conditioned on neural (local) shape attributes, enabling surface PDEs to be solved directly where the (neural) data lives. The operator integrates naturally with prevalent neural surface representations, is trained once on a single representative shape, and generalizes across shape and topology variations, enabling accurate, fast inference without explicit meshing or per-instance optimization while preserving differentiability. Across analytic benchmarks (heat equation and Poisson solve on sphere) and real neural assets across different representations, our method slightly outperforms CPM while remaining reasonably close to FEM, and, to our knowledge, delivers the first end-to-end pipeline that solves surface PDEs on both neural and classical surface representations. Code will be released on acceptance.
NegoCollab: A Common Representation Negotiation Approach for Heterogeneous Collaborative Perception
Oct 31, 2025Abstract:Collaborative perception improves task performance by expanding the perception range through information sharing among agents. . Immutable heterogeneity poses a significant challenge in collaborative perception, as participating agents may employ different and fixed perception models. This leads to domain gaps in the intermediate features shared among agents, consequently degrading collaborative performance. Aligning the features of all agents to a common representation can eliminate domain gaps with low training cost. However, in existing methods, the common representation is designated as the representation of a specific agent, making it difficult for agents with significant domain discrepancies from this specific agent to achieve proper alignment. This paper proposes NegoCollab, a heterogeneous collaboration method based on the negotiated common representation. It introduces a negotiator during training to derive the common representation from the local representations of each modality's agent, effectively reducing the inherent domain gap with the various local representations. In NegoCollab, the mutual transformation of features between the local representation space and the common representation space is achieved by a pair of sender and receiver. To better align local representations to the common representation containing multimodal information, we introduce structural alignment loss and pragmatic alignment loss in addition to the distribution alignment loss to supervise the training. This enables the knowledge in the common representation to be fully distilled into the sender.
CLR-Wire: Towards Continuous Latent Representations for 3D Curve Wireframe Generation
May 01, 2025Abstract:We introduce CLR-Wire, a novel framework for 3D curve-based wireframe generation that integrates geometry and topology into a unified Continuous Latent Representation. Unlike conventional methods that decouple vertices, edges, and faces, CLR-Wire encodes curves as Neural Parametric Curves along with their topological connectivity into a continuous and fixed-length latent space using an attention-driven variational autoencoder (VAE). This unified approach facilitates joint learning and generation of both geometry and topology. To generate wireframes, we employ a flow matching model to progressively map Gaussian noise to these latents, which are subsequently decoded into complete 3D wireframes. Our method provides fine-grained modeling of complex shapes and irregular topologies, and supports both unconditional generation and generation conditioned on point cloud or image inputs. Experimental results demonstrate that, compared with state-of-the-art generative approaches, our method achieves substantial improvements in accuracy, novelty, and diversity, offering an efficient and comprehensive solution for CAD design, geometric reconstruction, and 3D content creation.
HoLa: B-Rep Generation using a Holistic Latent Representation
Apr 22, 2025Abstract:We introduce a novel representation for learning and generating Computer-Aided Design (CAD) models in the form of $\textit{boundary representations}$ (B-Reps). Our representation unifies the continuous geometric properties of B-Rep primitives in different orders (e.g., surfaces and curves) and their discrete topological relations in a $\textit{holistic latent}$ (HoLa) space. This is based on the simple observation that the topological connection between two surfaces is intrinsically tied to the geometry of their intersecting curve. Such a prior allows us to reformulate topology learning in B-Reps as a geometric reconstruction problem in Euclidean space. Specifically, we eliminate the presence of curves, vertices, and all the topological connections in the latent space by learning to distinguish and derive curve geometries from a pair of surface primitives via a neural intersection network. To this end, our holistic latent space is only defined on surfaces but encodes a full B-Rep model, including the geometry of surfaces, curves, vertices, and their topological relations. Our compact and holistic latent space facilitates the design of a first diffusion-based generator to take on a large variety of inputs including point clouds, single/multi-view images, 2D sketches, and text prompts. Our method significantly reduces ambiguities, redundancies, and incoherences among the generated B-Rep primitives, as well as training complexities inherent in prior multi-step B-Rep learning pipelines, while achieving greatly improved validity rate over current state of the art: 82% vs. $\approx$50%.
Unicorn: A Universal and Collaborative Reinforcement Learning Approach Towards Generalizable Network-Wide Traffic Signal Control
Mar 14, 2025Abstract:Adaptive traffic signal control (ATSC) is crucial in reducing congestion, maximizing throughput, and improving mobility in rapidly growing urban areas. Recent advancements in parameter-sharing multi-agent reinforcement learning (MARL) have greatly enhanced the scalable and adaptive optimization of complex, dynamic flows in large-scale homogeneous networks. However, the inherent heterogeneity of real-world traffic networks, with their varied intersection topologies and interaction dynamics, poses substantial challenges to achieving scalable and effective ATSC across different traffic scenarios. To address these challenges, we present Unicorn, a universal and collaborative MARL framework designed for efficient and adaptable network-wide ATSC. Specifically, we first propose a unified approach to map the states and actions of intersections with varying topologies into a common structure based on traffic movements. Next, we design a Universal Traffic Representation (UTR) module with a decoder-only network for general feature extraction, enhancing the model's adaptability to diverse traffic scenarios. Additionally, we incorporate an Intersection Specifics Representation (ISR) module, designed to identify key latent vectors that represent the unique intersection's topology and traffic dynamics through variational inference techniques. To further refine these latent representations, we employ a contrastive learning approach in a self-supervised manner, which enables better differentiation of intersection-specific features. Moreover, we integrate the state-action dependencies of neighboring agents into policy optimization, which effectively captures dynamic agent interactions and facilitates efficient regional collaboration. Our results show that Unicorn outperforms other methods across various evaluation metrics, highlighting its potential in complex, dynamic traffic networks.
A Data-driven Dynamic Temporal Correlation Modeling Framework for Renewable Energy Scenario Generation
Jan 24, 2025Abstract:Renewable energy power is influenced by the atmospheric system, which exhibits nonlinear and time-varying features. To address this, a dynamic temporal correlation modeling framework is proposed for renewable energy scenario generation. A novel decoupled mapping path is employed for joint probability distribution modeling, formulating regression tasks for both marginal distributions and the correlation structure using proper scoring rules to ensure the rationality of the modeling process. The scenario generation process is divided into two stages. Firstly, the dynamic correlation network models temporal correlations based on a dynamic covariance matrix, capturing the time-varying features of renewable energy while enhancing the interpretability of the black-box model. Secondly, the implicit quantile network models the marginal quantile function in a nonparametric, continuous manner, enabling scenario generation through marginal inverse sampling. Experimental results demonstrate that the proposed dynamic correlation quantile network outperforms state-of-the-art methods in quantifying uncertainty and capturing dynamic correlation for short-term renewable energy scenario generation.
SinoSynth: A Physics-based Domain Randomization Approach for Generalizable CBCT Image Enhancement
Sep 27, 2024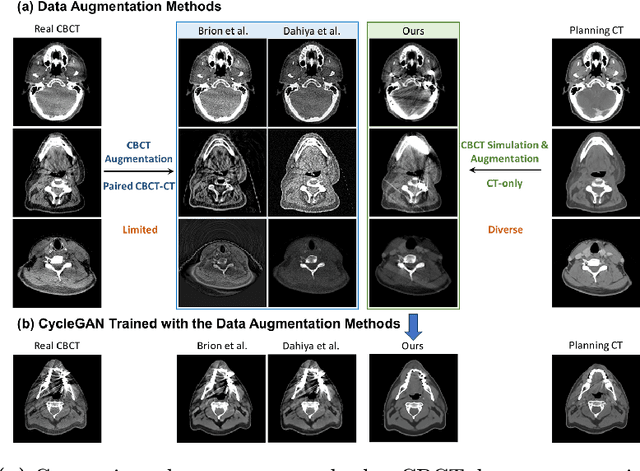
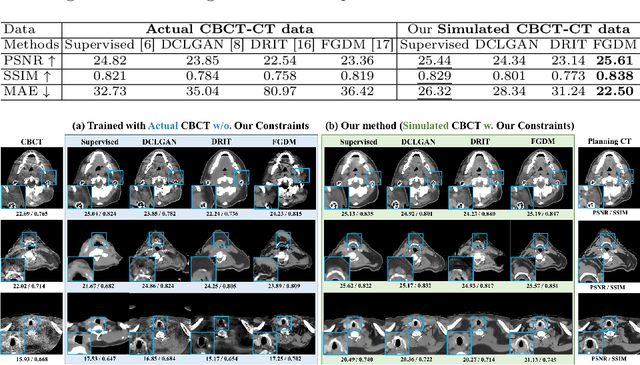
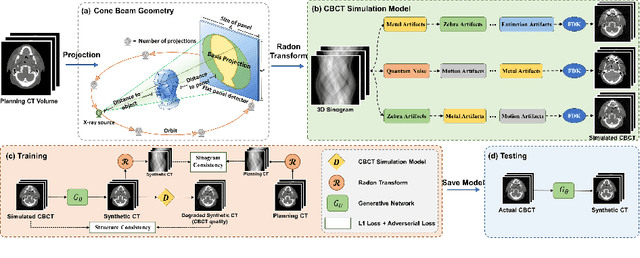
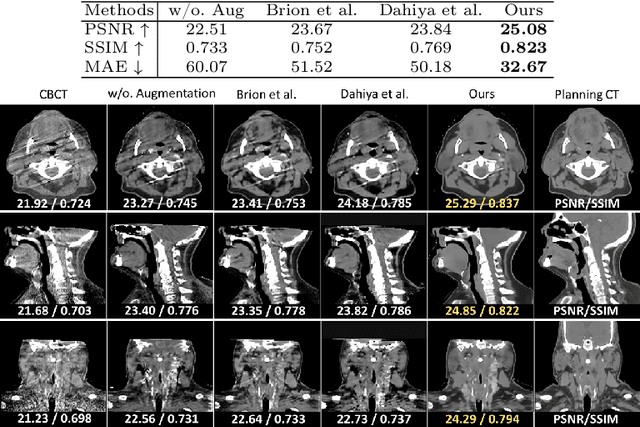
Abstract:Cone Beam Computed Tomography (CBCT) finds diverse applications in medicine. Ensuring high image quality in CBCT scans is essential for accurate diagnosis and treatment delivery. Yet, the susceptibility of CBCT images to noise and artifacts undermines both their usefulness and reliability. Existing methods typically address CBCT artifacts through image-to-image translation approaches. These methods, however, are limited by the artifact types present in the training data, which may not cover the complete spectrum of CBCT degradations stemming from variations in imaging protocols. Gathering additional data to encompass all possible scenarios can often pose a challenge. To address this, we present SinoSynth, a physics-based degradation model that simulates various CBCT-specific artifacts to generate a diverse set of synthetic CBCT images from high-quality CT images without requiring pre-aligned data. Through extensive experiments, we demonstrate that several different generative networks trained on our synthesized data achieve remarkable results on heterogeneous multi-institutional datasets, outperforming even the same networks trained on actual data. We further show that our degradation model conveniently provides an avenue to enforce anatomical constraints in conditional generative models, yielding high-quality and structure-preserving synthetic CT images.
Generating 3D House Wireframes with Semantics
Jul 17, 2024Abstract:We present a new approach for generating 3D house wireframes with semantic enrichment using an autoregressive model. Unlike conventional generative models that independently process vertices, edges, and faces, our approach employs a unified wire-based representation for improved coherence in learning 3D wireframe structures. By re-ordering wire sequences based on semantic meanings, we facilitate seamless semantic integration during sequence generation. Our two-phase technique merges a graph-based autoencoder with a transformer-based decoder to learn latent geometric tokens and generate semantic-aware wireframes. Through iterative prediction and decoding during inference, our model produces detailed wireframes that can be easily segmented into distinct components, such as walls, roofs, and rooms, reflecting the semantic essence of the shape. Empirical results on a comprehensive house dataset validate the superior accuracy, novelty, and semantic fidelity of our model compared to existing generative models. More results and details can be found on https://vcc.tech/research/2024/3DWire.
Split-and-Fit: Learning B-Reps via Structure-Aware Voronoi Partitioning
Jun 07, 2024



Abstract:We introduce a novel method for acquiring boundary representations (B-Reps) of 3D CAD models which involves a two-step process: it first applies a spatial partitioning, referred to as the ``split``, followed by a ``fit`` operation to derive a single primitive within each partition. Specifically, our partitioning aims to produce the classical Voronoi diagram of the set of ground-truth (GT) B-Rep primitives. In contrast to prior B-Rep constructions which were bottom-up, either via direct primitive fitting or point clustering, our Split-and-Fit approach is top-down and structure-aware, since a Voronoi partition explicitly reveals both the number of and the connections between the primitives. We design a neural network to predict the Voronoi diagram from an input point cloud or distance field via a binary classification. We show that our network, coined NVD-Net for neural Voronoi diagrams, can effectively learn Voronoi partitions for CAD models from training data and exhibits superior generalization capabilities. Extensive experiments and evaluation demonstrate that the resulting B-Reps, consisting of parametric surfaces, curves, and vertices, are more plausible than those obtained by existing alternatives, with significant improvements in reconstruction quality. Code will be released on https://github.com/yilinliu77/NVDNet.
CSANet: Channel Spatial Attention Network for Robust 3D Face Alignment and Reconstruction
May 30, 2024



Abstract:Our project proposes an end-to-end 3D face alignment and reconstruction network. The backbone of our model is built by Bottle-Neck structure via Depth-wise Separable Convolution. We integrate Coordinate Attention mechanism and Spatial Group-wise Enhancement to extract more representative features. For more stable training process and better convergence, we jointly use Wing loss and the Weighted Parameter Distance Cost to learn parameters for 3D Morphable model and 3D vertices. Our proposed model outperforms all baseline models both quantitatively and qualitatively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge