Ling Yue
Foam-Agent: Towards Automated Intelligent CFD Workflows
May 08, 2025Abstract:Computational Fluid Dynamics (CFD) is an essential simulation tool in various engineering disciplines, but it often requires substantial domain expertise and manual configuration, creating barriers to entry. We present Foam-Agent, a multi-agent framework that automates complex OpenFOAM-based CFD simulation workflows from natural language inputs. Our innovation includes (1) a hierarchical multi-index retrieval system with specialized indices for different simulation aspects, (2) a dependency-aware file generation system that provides consistency management across configuration files, and (3) an iterative error correction mechanism that diagnoses and resolves simulation failures without human intervention. Through comprehensive evaluation on the dataset of 110 simulation tasks, Foam-Agent achieves an 83.6% success rate with Claude 3.5 Sonnet, significantly outperforming existing frameworks (55.5% for MetaOpenFOAM and 37.3% for OpenFOAM-GPT). Ablation studies demonstrate the critical contribution of each system component, with the specialized error correction mechanism providing a 36.4% performance improvement. Foam-Agent substantially lowers the CFD expertise threshold while maintaining modeling accuracy, demonstrating the potential of specialized multi-agent systems to democratize access to complex scientific simulation tools. The code is public at https://github.com/csml-rpi/Foam-Agent
The Potential and Value of AI Chatbot in Personalized Cognitive Training
Oct 25, 2024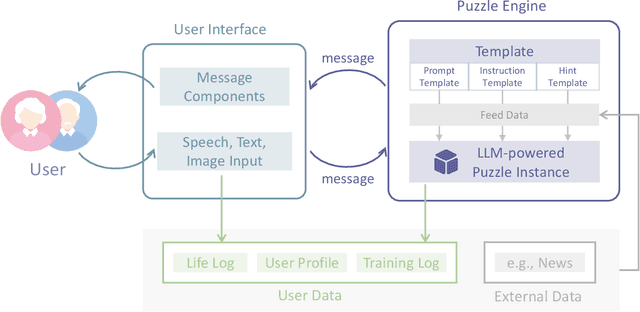


Abstract:In recent years, the rapid aging of the global population has led to an increase in cognitive disorders, such as Alzheimer's disease, presenting significant public health challenges. Although no effective treatments currently exist to reverse Alzheimer's, prevention and early intervention, including cognitive training, are critical. This report explores the potential of AI chatbots in enhancing personalized cognitive training. We introduce ReMe, a web-based framework designed to create AI chatbots that facilitate cognitive training research, specifically targeting episodic memory tasks derived from personal life logs. By leveraging large language models, ReMe provides enhanced user-friendly, interactive, and personalized training experiences. Case studies demonstrate ReMe's effectiveness in engaging users through life recall and open-ended language puzzles, highlighting its potential to improve cognitive training design. Despite promising results, further research is needed to validate training effectiveness through large-scale studies that include cognitive ability evaluations. Overall, ReMe offers a promising approach to personalized cognitive training, utilizing AI capabilities to meet the growing demand for non-pharmacological interventions in cognitive health, with future research aiming to expand its applications and efficacy.
SMILES-Mamba: Chemical Mamba Foundation Models for Drug ADMET Prediction
Aug 11, 2024
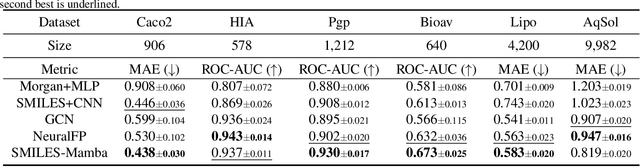


Abstract:In drug discovery, predicting the absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of small-molecule drugs is critical for ensuring safety and efficacy. However, the process of accurately predicting these properties is often resource-intensive and requires extensive experimental data. To address this challenge, we propose SMILES-Mamba, a two-stage model that leverages both unlabeled and labeled data through a combination of self-supervised pretraining and fine-tuning strategies. The model first pre-trains on a large corpus of unlabeled SMILES strings to capture the underlying chemical structure and relationships, before being fine-tuned on smaller, labeled datasets specific to ADMET tasks. Our results demonstrate that SMILES-Mamba exhibits competitive performance across 22 ADMET datasets, achieving the highest score in 14 tasks, highlighting the potential of self-supervised learning in improving molecular property prediction. This approach not only enhances prediction accuracy but also reduces the dependence on large, labeled datasets, offering a promising direction for future research in drug discovery.
BioMamba: A Pre-trained Biomedical Language Representation Model Leveraging Mamba
Aug 05, 2024
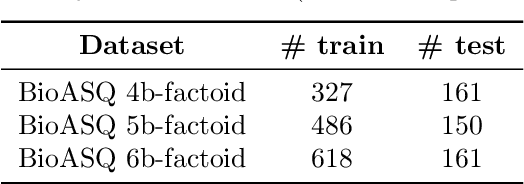
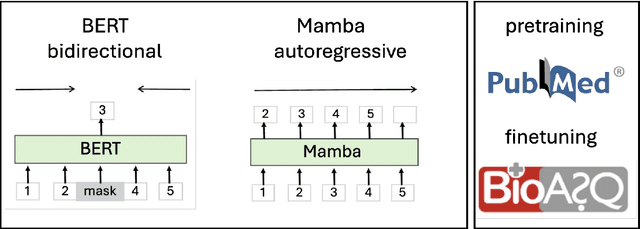
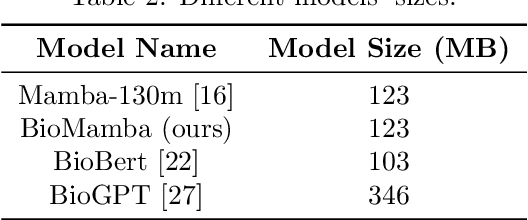
Abstract:The advancement of natural language processing (NLP) in biology hinges on models' ability to interpret intricate biomedical literature. Traditional models often struggle with the complex and domain-specific language in this field. In this paper, we present BioMamba, a pre-trained model specifically designed for biomedical text mining. BioMamba builds upon the Mamba architecture and is pre-trained on an extensive corpus of biomedical literature. Our empirical studies demonstrate that BioMamba significantly outperforms models like BioBERT and general-domain Mamba across various biomedical tasks. For instance, BioMamba achieves a 100 times reduction in perplexity and a 4 times reduction in cross-entropy loss on the BioASQ test set. We provide an overview of the model architecture, pre-training process, and fine-tuning techniques. Additionally, we release the code and trained model to facilitate further research.
TrialEnroll: Predicting Clinical Trial Enrollment Success with Deep & Cross Network and Large Language Models
Jul 18, 2024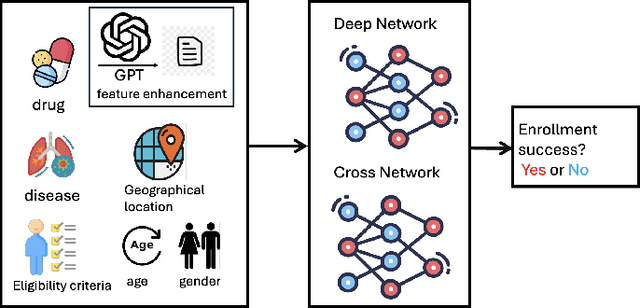

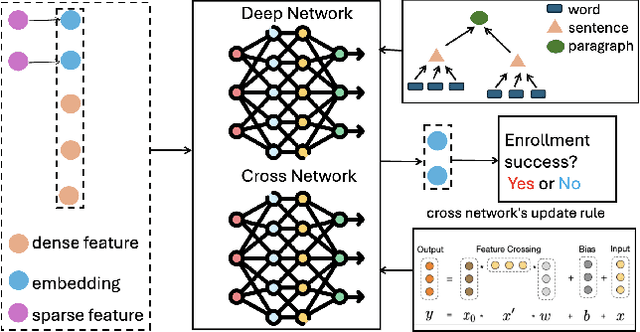
Abstract:Clinical trials need to recruit a sufficient number of volunteer patients to demonstrate the statistical power of the treatment (e.g., a new drug) in curing a certain disease. Clinical trial recruitment has a significant impact on trial success. Forecasting whether the recruitment process would be successful before we run the trial would save many resources and time. This paper develops a novel deep & cross network with large language model (LLM)-augmented text feature that learns semantic information from trial eligibility criteria and predicts enrollment success. The proposed method enables interpretability by understanding which sentence/word in eligibility criteria contributes heavily to prediction. We also demonstrate the empirical superiority of the proposed method (0.7002 PR-AUC) over a bunch of well-established machine learning methods. The code and curated dataset are publicly available at https://anonymous.4open.science/r/TrialEnroll-7E12.
Functional Imaging Constrained Diffusion for Brain PET Synthesis from Structural MRI
May 03, 2024



Abstract:Magnetic resonance imaging (MRI) and positron emission tomography (PET) are increasingly used in multimodal analysis of neurodegenerative disorders. While MRI is broadly utilized in clinical settings, PET is less accessible. Many studies have attempted to use deep generative models to synthesize PET from MRI scans. However, they often suffer from unstable training and inadequately preserve brain functional information conveyed by PET. To this end, we propose a functional imaging constrained diffusion (FICD) framework for 3D brain PET image synthesis with paired structural MRI as input condition, through a new constrained diffusion model (CDM). The FICD introduces noise to PET and then progressively removes it with CDM, ensuring high output fidelity throughout a stable training phase. The CDM learns to predict denoised PET with a functional imaging constraint introduced to ensure voxel-wise alignment between each denoised PET and its ground truth. Quantitative and qualitative analyses conducted on 293 subjects with paired T1-weighted MRI and 18F-fluorodeoxyglucose (FDG)-PET scans suggest that FICD achieves superior performance in generating FDG-PET data compared to state-of-the-art methods. We further validate the effectiveness of the proposed FICD on data from a total of 1,262 subjects through three downstream tasks, with experimental results suggesting its utility and generalizability.
CT-Agent: Clinical Trial Multi-Agent with Large Language Model-based Reasoning
Apr 23, 2024Abstract:Large Language Models (LLMs) and multi-agent systems have shown impressive capabilities in natural language tasks but face challenges in clinical trial applications, primarily due to limited access to external knowledge. Recognizing the potential of advanced clinical trial tools that aggregate and predict based on the latest medical data, we propose an integrated solution to enhance their accessibility and utility. We introduce Clinical Agent System (CT-Agent), a Clinical multi-agent system designed for clinical trial tasks, leveraging GPT-4, multi-agent architectures, LEAST-TO-MOST, and ReAct reasoning technology. This integration not only boosts LLM performance in clinical contexts but also introduces novel functionalities. Our system autonomously manages the entire clinical trial process, demonstrating significant efficiency improvements in our evaluations, which include both computational benchmarks and expert feedback.
TrialDura: Hierarchical Attention Transformer for Interpretable Clinical Trial Duration Prediction
Apr 20, 2024



Abstract:The clinical trial process, also known as drug development, is an indispensable step toward the development of new treatments. The major objective of interventional clinical trials is to assess the safety and effectiveness of drug-based treatment in treating certain diseases in the human body. However, clinical trials are lengthy, labor-intensive, and costly. The duration of a clinical trial is a crucial factor that influences overall expenses. Therefore, effective management of the timeline of a clinical trial is essential for controlling the budget and maximizing the economic viability of the research. To address this issue, We propose TrialDura, a machine learning-based method that estimates the duration of clinical trials using multimodal data, including disease names, drug molecules, trial phases, and eligibility criteria. Then, we encode them into Bio-BERT embeddings specifically tuned for biomedical contexts to provide a deeper and more relevant semantic understanding of clinical trial data. Finally, the model's hierarchical attention mechanism connects all of the embeddings to capture their interactions and predict clinical trial duration. Our proposed model demonstrated superior performance with a mean absolute error (MAE) of 1.04 years and a root mean square error (RMSE) of 1.39 years compared to the other models, indicating more accurate clinical trial duration prediction. Publicly available code can be found at https://anonymous.4open.science/r/TrialDura-F196
Emerging Drug Interaction Prediction Enabled by Flow-based Graph Neural Network with Biomedical Network
Nov 15, 2023



Abstract:Accurately predicting drug-drug interactions (DDI) for emerging drugs, which offer possibilities for treating and alleviating diseases, with computational methods can improve patient care and contribute to efficient drug development. However, many existing computational methods require large amounts of known DDI information, which is scarce for emerging drugs. In this paper, we propose EmerGNN, a graph neural network (GNN) that can effectively predict interactions for emerging drugs by leveraging the rich information in biomedical networks. EmerGNN learns pairwise representations of drugs by extracting the paths between drug pairs, propagating information from one drug to the other, and incorporating the relevant biomedical concepts on the paths. The different edges on the biomedical network are weighted to indicate the relevance for the target DDI prediction. Overall, EmerGNN has higher accuracy than existing approaches in predicting interactions for emerging drugs and can identify the most relevant information on the biomedical network.
Ensemble Learning for Graph Neural Networks
Oct 22, 2023Abstract:Graph Neural Networks (GNNs) have shown success in various fields for learning from graph-structured data. This paper investigates the application of ensemble learning techniques to improve the performance and robustness of Graph Neural Networks (GNNs). By training multiple GNN models with diverse initializations or architectures, we create an ensemble model named ELGNN that captures various aspects of the data and uses the Tree-Structured Parzen Estimator algorithm to determine the ensemble weights. Combining the predictions of these models enhances overall accuracy, reduces bias and variance, and mitigates the impact of noisy data. Our findings demonstrate the efficacy of ensemble learning in enhancing GNN capabilities for analyzing complex graph-structured data. The code is public at https://github.com/wongzhenhao/ELGNN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge