Chenhao Li
Scaling Rough Terrain Locomotion with Automatic Curriculum Reinforcement Learning
Jan 24, 2026Abstract:Curriculum learning has demonstrated substantial effectiveness in robot learning. However, it still faces limitations when scaling to complex, wide-ranging task spaces. Such task spaces often lack a well-defined difficulty structure, making the difficulty ordering required by previous methods challenging to define. We propose a Learning Progress-based Automatic Curriculum Reinforcement Learning (LP-ACRL) framework, which estimates the agent's learning progress online and adaptively adjusts the task-sampling distribution, thereby enabling automatic curriculum generation without prior knowledge of the difficulty distribution over the task space. Policies trained with LP-ACRL enable the ANYmal D quadruped to achieve and maintain stable, high-speed locomotion at 2.5 m/s linear velocity and 3.0 rad/s angular velocity across diverse terrains, including stairs, slopes, gravel, and low-friction flat surfaces--whereas previous methods have generally been limited to high speeds on flat terrain or low speeds on complex terrain. Experimental results demonstrate that LP-ACRL exhibits strong scalability and real-world applicability, providing a robust baseline for future research on curriculum generation in complex, wide-ranging robotic learning task spaces.
ActiShade: Activating Overshadowed Knowledge to Guide Multi-Hop Reasoning in Large Language Models
Jan 12, 2026Abstract:In multi-hop reasoning, multi-round retrieval-augmented generation (RAG) methods typically rely on LLM-generated content as the retrieval query. However, these approaches are inherently vulnerable to knowledge overshadowing - a phenomenon where critical information is overshadowed during generation. As a result, the LLM-generated content may be incomplete or inaccurate, leading to irrelevant retrieval and causing error accumulation during the iteration process. To address this challenge, we propose ActiShade, which detects and activates overshadowed knowledge to guide large language models (LLMs) in multi-hop reasoning. Specifically, ActiShade iteratively detects the overshadowed keyphrase in the given query, retrieves documents relevant to both the query and the overshadowed keyphrase, and generates a new query based on the retrieved documents to guide the next-round iteration. By supplementing the overshadowed knowledge during the formulation of next-round queries while minimizing the introduction of irrelevant noise, ActiShade reduces the error accumulation caused by knowledge overshadowing. Extensive experiments show that ActiShade outperforms existing methods across multiple datasets and LLMs.
Multi-Domain Motion Embedding: Expressive Real-Time Mimicry for Legged Robots
Dec 08, 2025



Abstract:Effective motion representation is crucial for enabling robots to imitate expressive behaviors in real time, yet existing motion controllers often ignore inherent patterns in motion. Previous efforts in representation learning do not attempt to jointly capture structured periodic patterns and irregular variations in human and animal movement. To address this, we present Multi-Domain Motion Embedding (MDME), a motion representation that unifies the embedding of structured and unstructured features using a wavelet-based encoder and a probabilistic embedding in parallel. This produces a rich representation of reference motions from a minimal input set, enabling improved generalization across diverse motion styles and morphologies. We evaluate MDME on retargeting-free real-time motion imitation by conditioning robot control policies on the learned embeddings, demonstrating accurate reproduction of complex trajectories on both humanoid and quadruped platforms. Our comparative studies confirm that MDME outperforms prior approaches in reconstruction fidelity and generalizability to unseen motions. Furthermore, we demonstrate that MDME can reproduce novel motion styles in real-time through zero-shot deployment, eliminating the need for task-specific tuning or online retargeting. These results position MDME as a generalizable and structure-aware foundation for scalable real-time robot imitation.
Learning Soft Robotic Dynamics with Active Exploration
Oct 31, 2025Abstract:Soft robots offer unmatched adaptability and safety in unstructured environments, yet their compliant, high-dimensional, and nonlinear dynamics make modeling for control notoriously difficult. Existing data-driven approaches often fail to generalize, constrained by narrowly focused task demonstrations or inefficient random exploration. We introduce SoftAE, an uncertainty-aware active exploration framework that autonomously learns task-agnostic and generalizable dynamics models of soft robotic systems. SoftAE employs probabilistic ensemble models to estimate epistemic uncertainty and actively guides exploration toward underrepresented regions of the state-action space, achieving efficient coverage of diverse behaviors without task-specific supervision. We evaluate SoftAE on three simulated soft robotic platforms -- a continuum arm, an articulated fish in fluid, and a musculoskeletal leg with hybrid actuation -- and on a pneumatically actuated continuum soft arm in the real world. Compared with random exploration and task-specific model-based reinforcement learning, SoftAE produces more accurate dynamics models, enables superior zero-shot control on unseen tasks, and maintains robustness under sensing noise, actuation delays, and nonlinear material effects. These results demonstrate that uncertainty-driven active exploration can yield scalable, reusable dynamics models across diverse soft robotic morphologies, representing a step toward more autonomous, adaptable, and data-efficient control in compliant robots.
Motion Priors Reimagined: Adapting Flat-Terrain Skills for Complex Quadruped Mobility
May 21, 2025Abstract:Reinforcement learning (RL)-based legged locomotion controllers often require meticulous reward tuning to track velocities or goal positions while preserving smooth motion on various terrains. Motion imitation methods via RL using demonstration data reduce reward engineering but fail to generalize to novel environments. We address this by proposing a hierarchical RL framework in which a low-level policy is first pre-trained to imitate animal motions on flat ground, thereby establishing motion priors. A subsequent high-level, goal-conditioned policy then builds on these priors, learning residual corrections that enable perceptive locomotion, local obstacle avoidance, and goal-directed navigation across diverse and rugged terrains. Simulation experiments illustrate the effectiveness of learned residuals in adapting to progressively challenging uneven terrains while still preserving the locomotion characteristics provided by the motion priors. Furthermore, our results demonstrate improvements in motion regularization over baseline models trained without motion priors under similar reward setups. Real-world experiments with an ANYmal-D quadruped robot confirm our policy's capability to generalize animal-like locomotion skills to complex terrains, demonstrating smooth and efficient locomotion and local navigation performance amidst challenging terrains with obstacles.
Offline Robotic World Model: Learning Robotic Policies without a Physics Simulator
Apr 23, 2025Abstract:Reinforcement Learning (RL) has demonstrated impressive capabilities in robotic control but remains challenging due to high sample complexity, safety concerns, and the sim-to-real gap. While offline RL eliminates the need for risky real-world exploration by learning from pre-collected data, it suffers from distributional shift, limiting policy generalization. Model-Based RL (MBRL) addresses this by leveraging predictive models for synthetic rollouts, yet existing approaches often lack robust uncertainty estimation, leading to compounding errors in offline settings. We introduce Offline Robotic World Model (RWM-O), a model-based approach that explicitly estimates epistemic uncertainty to improve policy learning without reliance on a physics simulator. By integrating these uncertainty estimates into policy optimization, our approach penalizes unreliable transitions, reducing overfitting to model errors and enhancing stability. Experimental results show that RWM-O improves generalization and safety, enabling policy learning purely from real-world data and advancing scalable, data-efficient RL for robotics.
NIL: No-data Imitation Learning by Leveraging Pre-trained Video Diffusion Models
Mar 13, 2025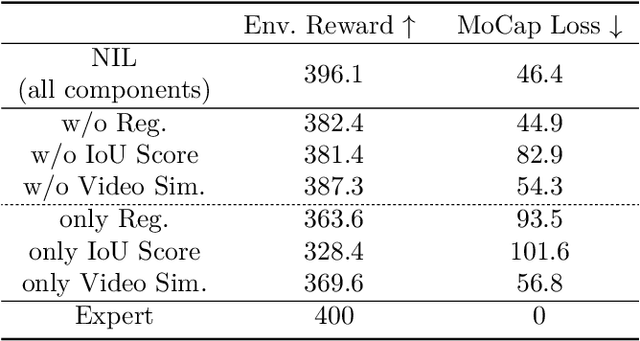
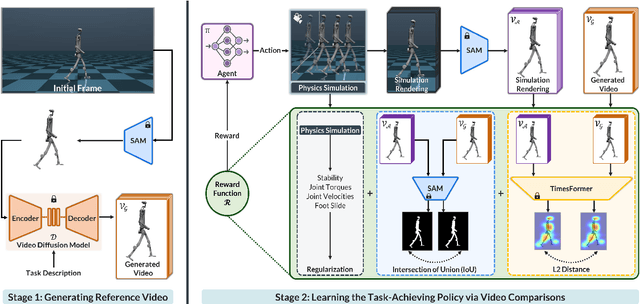
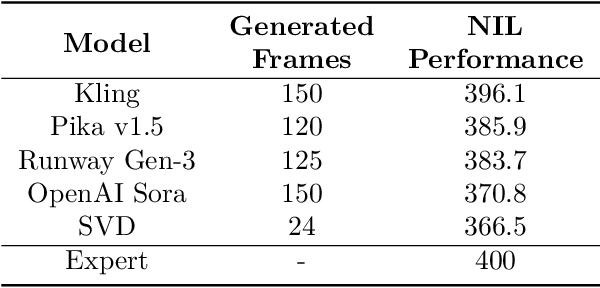
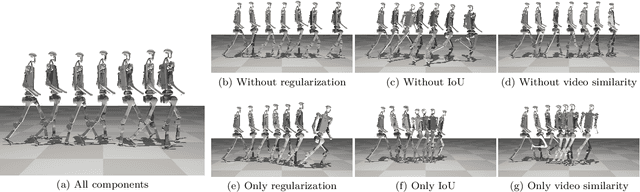
Abstract:Acquiring physically plausible motor skills across diverse and unconventional morphologies-including humanoid robots, quadrupeds, and animals-is essential for advancing character simulation and robotics. Traditional methods, such as reinforcement learning (RL) are task- and body-specific, require extensive reward function engineering, and do not generalize well. Imitation learning offers an alternative but relies heavily on high-quality expert demonstrations, which are difficult to obtain for non-human morphologies. Video diffusion models, on the other hand, are capable of generating realistic videos of various morphologies, from humans to ants. Leveraging this capability, we propose a data-independent approach for skill acquisition that learns 3D motor skills from 2D-generated videos, with generalization capability to unconventional and non-human forms. Specifically, we guide the imitation learning process by leveraging vision transformers for video-based comparisons by calculating pair-wise distance between video embeddings. Along with video-encoding distance, we also use a computed similarity between segmented video frames as a guidance reward. We validate our method on locomotion tasks involving unique body configurations. In humanoid robot locomotion tasks, we demonstrate that 'No-data Imitation Learning' (NIL) outperforms baselines trained on 3D motion-capture data. Our results highlight the potential of leveraging generative video models for physically plausible skill learning with diverse morphologies, effectively replacing data collection with data generation for imitation learning.
Toward Task Generalization via Memory Augmentation in Meta-Reinforcement Learning
Feb 03, 2025



Abstract:In reinforcement learning (RL), agents often struggle to perform well on tasks that differ from those encountered during training. This limitation presents a challenge to the broader deployment of RL in diverse and dynamic task settings. In this work, we introduce memory augmentation, a memory-based RL approach to improve task generalization. Our approach leverages task-structured augmentations to simulate plausible out-of-distribution scenarios and incorporates memory mechanisms to enable context-aware policy adaptation. Trained on a predefined set of tasks, our policy demonstrates the ability to generalize to unseen tasks through memory augmentation without requiring additional interactions with the environment. Through extensive simulation experiments and real-world hardware evaluations on legged locomotion tasks, we demonstrate that our approach achieves zero-shot generalization to unseen tasks while maintaining robust in-distribution performance and high sample efficiency.
Learning More With Less: Sample Efficient Dynamics Learning and Model-Based RL for Loco-Manipulation
Jan 17, 2025



Abstract:Combining the agility of legged locomotion with the capabilities of manipulation, loco-manipulation platforms have the potential to perform complex tasks in real-world applications. To this end, state-of-the-art quadrupeds with attached manipulators, such as the Boston Dynamics Spot, have emerged to provide a capable and robust platform. However, both the complexity of loco-manipulation control, as well as the black-box nature of commercial platforms pose challenges for developing accurate dynamics models and control policies. We address these challenges by developing a hand-crafted kinematic model for a quadruped-with-arm platform and, together with recent advances in Bayesian Neural Network (BNN)-based dynamics learning using physical priors, efficiently learn an accurate dynamics model from data. We then derive control policies for loco-manipulation via model-based reinforcement learning (RL). We demonstrate the effectiveness of this approach on hardware using the Boston Dynamics Spot with a manipulator, accurately performing dynamic end-effector trajectory tracking even in low data regimes.
Robotic World Model: A Neural Network Simulator for Robust Policy Optimization in Robotics
Jan 17, 2025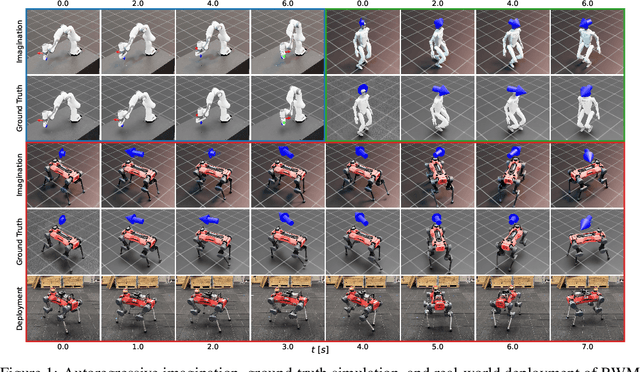
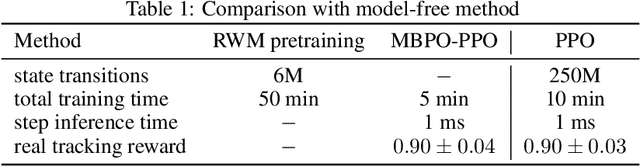
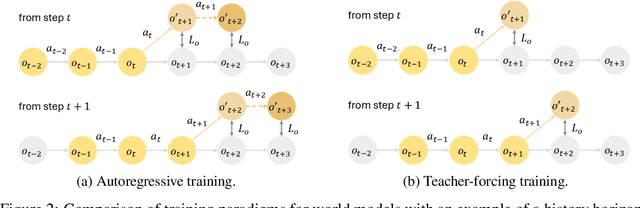
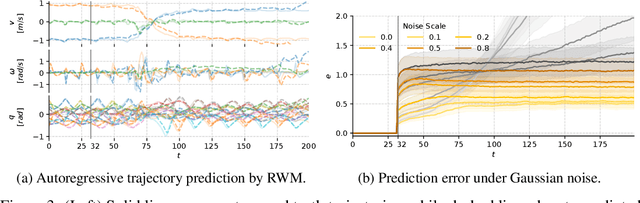
Abstract:Learning robust and generalizable world models is crucial for enabling efficient and scalable robotic control in real-world environments. In this work, we introduce a novel framework for learning world models that accurately capture complex, partially observable, and stochastic dynamics. The proposed method employs a dual-autoregressive mechanism and self-supervised training to achieve reliable long-horizon predictions without relying on domain-specific inductive biases, ensuring adaptability across diverse robotic tasks. We further propose a policy optimization framework that leverages world models for efficient training in imagined environments and seamless deployment in real-world systems. Through extensive experiments, our approach consistently outperforms state-of-the-art methods, demonstrating superior autoregressive prediction accuracy, robustness to noise, and generalization across manipulation and locomotion tasks. Notably, policies trained with our method are successfully deployed on ANYmal D hardware in a zero-shot transfer, achieving robust performance with minimal sim-to-real performance loss. This work advances model-based reinforcement learning by addressing the challenges of long-horizon prediction, error accumulation, and sim-to-real transfer. By providing a scalable and robust framework, the introduced methods pave the way for adaptive and efficient robotic systems in real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge