Yongqi Zhang
Mixture of Length and Pruning Experts for Knowledge Graphs Reasoning
Jul 28, 2025Abstract:Knowledge Graph (KG) reasoning, which aims to infer new facts from structured knowledge repositories, plays a vital role in Natural Language Processing (NLP) systems. Its effectiveness critically depends on constructing informative and contextually relevant reasoning paths. However, existing graph neural networks (GNNs) often adopt rigid, query-agnostic path-exploration strategies, limiting their ability to adapt to diverse linguistic contexts and semantic nuances. To address these limitations, we propose \textbf{MoKGR}, a mixture-of-experts framework that personalizes path exploration through two complementary components: (1) a mixture of length experts that adaptively selects and weights candidate path lengths according to query complexity, providing query-specific reasoning depth; and (2) a mixture of pruning experts that evaluates candidate paths from a complementary perspective, retaining the most informative paths for each query. Through comprehensive experiments on diverse benchmark, MoKGR demonstrates superior performance in both transductive and inductive settings, validating the effectiveness of personalized path exploration in KGs reasoning.
Case-Based Reasoning Enhances the Predictive Power of LLMs in Drug-Drug Interaction
May 29, 2025Abstract:Drug-drug interaction (DDI) prediction is critical for treatment safety. While large language models (LLMs) show promise in pharmaceutical tasks, their effectiveness in DDI prediction remains challenging. Inspired by the well-established clinical practice where physicians routinely reference similar historical cases to guide their decisions through case-based reasoning (CBR), we propose CBR-DDI, a novel framework that distills pharmacological principles from historical cases to improve LLM reasoning for DDI tasks. CBR-DDI constructs a knowledge repository by leveraging LLMs to extract pharmacological insights and graph neural networks (GNNs) to model drug associations. A hybrid retrieval mechanism and dual-layer knowledge-enhanced prompting allow LLMs to effectively retrieve and reuse relevant cases. We further introduce a representative sampling strategy for dynamic case refinement. Extensive experiments demonstrate that CBR-DDI achieves state-of-the-art performance, with a significant 28.7% accuracy improvement over both popular LLMs and CBR baseline, while maintaining high interpretability and flexibility.
GraphOracle: A Foundation Model for Knowledge Graph Reasoning
May 16, 2025Abstract:Foundation models have demonstrated remarkable capabilities across various domains, but developing analogous models for knowledge graphs presents unique challenges due to their dynamic nature and the need for cross-domain reasoning. To address these issues, we introduce \textbf{\textsc{GraphOracle}}, a relation-centric foundation model that unifies reasoning across knowledge graphs by converting them into Relation-Dependency Graphs (RDG), explicitly encoding compositional patterns with fewer edges than prior methods. A query-dependent attention mechanism is further developed to learn inductive representations for both relations and entities. Pre-training on diverse knowledge graphs, followed by minutes-level fine-tuning, enables effective generalization to unseen entities, relations, and entire graphs. Through comprehensive experiments on 31 diverse benchmarks spanning transductive, inductive, and cross-domain settings, we demonstrate consistent state-of-the-art performance with minimal adaptation, improving the prediction performance by up to 35\% compared to the strongest baselines.
An LLM-enabled Multi-Agent Autonomous Mechatronics Design Framework
Apr 20, 2025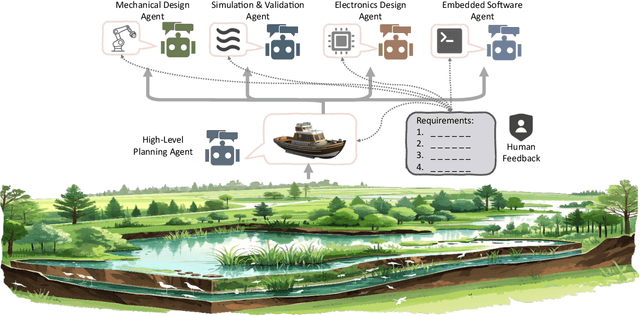

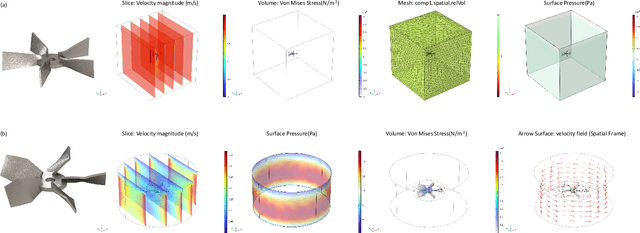
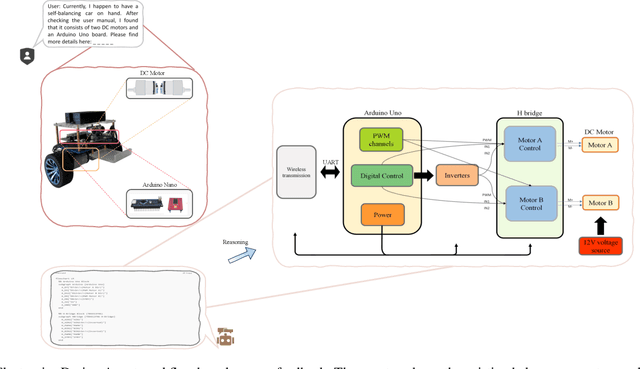
Abstract:Existing LLM-enabled multi-agent frameworks are predominantly limited to digital or simulated environments and confined to narrowly focused knowledge domain, constraining their applicability to complex engineering tasks that require the design of physical embodiment, cross-disciplinary integration, and constraint-aware reasoning. This work proposes a multi-agent autonomous mechatronics design framework, integrating expertise across mechanical design, optimization, electronics, and software engineering to autonomously generate functional prototypes with minimal direct human design input. Operating primarily through a language-driven workflow, the framework incorporates structured human feedback to ensure robust performance under real-world constraints. To validate its capabilities, the framework is applied to a real-world challenge involving autonomous water-quality monitoring and sampling, where traditional methods are labor-intensive and ecologically disruptive. Leveraging the proposed system, a fully functional autonomous vessel was developed with optimized propulsion, cost-effective electronics, and advanced control. The design process was carried out by specialized agents, including a high-level planning agent responsible for problem abstraction and dedicated agents for structural, electronics, control, and software development. This approach demonstrates the potential of LLM-based multi-agent systems to automate real-world engineering workflows and reduce reliance on extensive domain expertise.
Activation-aware Probe-Query: Effective Key-Value Retrieval for Long-Context LLMs Inference
Feb 19, 2025Abstract:Recent advances in large language models (LLMs) have showcased exceptional performance in long-context tasks, while facing significant inference efficiency challenges with limited GPU memory. Existing solutions first proposed the sliding-window approach to accumulate a set of historical \textbf{key-value} (KV) pairs for reuse, then further improvements selectively retain its subsets at each step. However, due to the sparse attention distribution across a long context, it is hard to identify and recall relevant KV pairs, as the attention is distracted by massive candidate pairs. Additionally, we found it promising to select representative tokens as probe-Query in each sliding window to effectively represent the entire context, which is an approach overlooked by existing methods. Thus, we propose \textbf{ActQKV}, a training-free, \textbf{Act}ivation-aware approach that dynamically determines probe-\textbf{Q}uery and leverages it to retrieve the relevant \textbf{KV} pairs for inference. Specifically, ActQKV monitors a token-level indicator, Activation Bias, within each context window, enabling the proper construction of probe-Query for retrieval at pre-filling stage. To accurately recall the relevant KV pairs and minimize the irrelevant ones, we design a dynamic KV cut-off mechanism guided by information density across layers at the decoding stage. Experiments on the Long-Bench and $\infty$ Benchmarks demonstrate its state-of-the-art performance with competitive inference quality and resource efficiency.
Perovskite-LLM: Knowledge-Enhanced Large Language Models for Perovskite Solar Cell Research
Feb 18, 2025Abstract:The rapid advancement of perovskite solar cells (PSCs) has led to an exponential growth in research publications, creating an urgent need for efficient knowledge management and reasoning systems in this domain. We present a comprehensive knowledge-enhanced system for PSCs that integrates three key components. First, we develop Perovskite-KG, a domain-specific knowledge graph constructed from 1,517 research papers, containing 23,789 entities and 22,272 relationships. Second, we create two complementary datasets: Perovskite-Chat, comprising 55,101 high-quality question-answer pairs generated through a novel multi-agent framework, and Perovskite-Reasoning, containing 2,217 carefully curated materials science problems. Third, we introduce two specialized large language models: Perovskite-Chat-LLM for domain-specific knowledge assistance and Perovskite-Reasoning-LLM for scientific reasoning tasks. Experimental results demonstrate that our system significantly outperforms existing models in both domain-specific knowledge retrieval and scientific reasoning tasks, providing researchers with effective tools for literature review, experimental design, and complex problem-solving in PSC research.
High-Sensitivity Vision-Based Tactile Sensing Enhanced by Microstructures and Lightweight CNN
Dec 30, 2024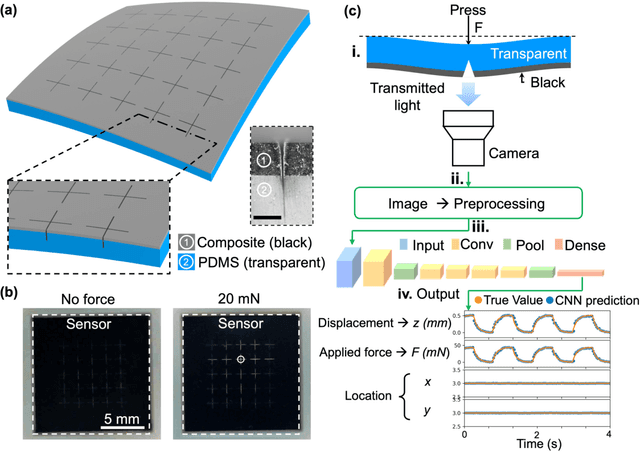
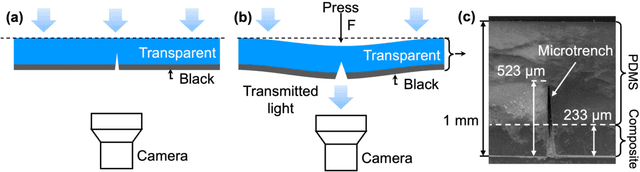
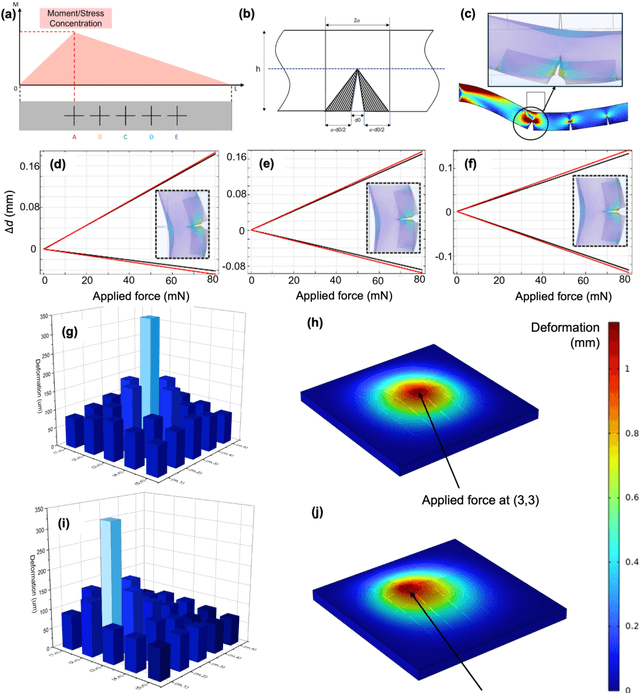
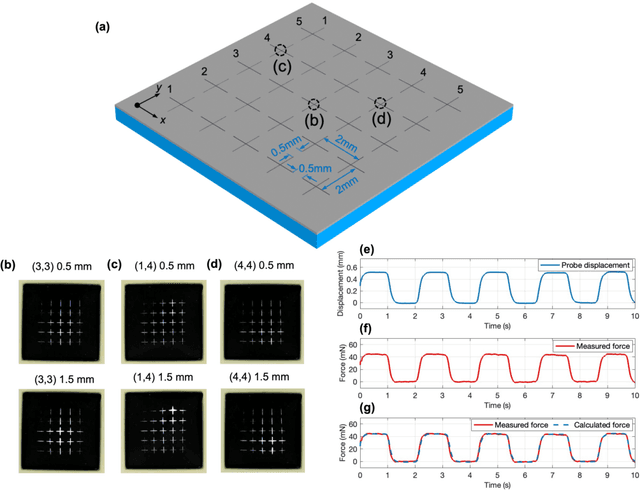
Abstract:Tactile sensing is critical in advanced interactive systems by emulating the human sense of touch to detect stimuli. Vision-based tactile sensors (VBTSs) are promising for their ability to provide rich information, robustness, adaptability, low cost, and multimodal capabilities. However, current technologies still have limitations in sensitivity, spatial resolution, and the high computational demands of deep learning-based image processing. This paper presents a comprehensive approach combining a novel sensor structure with micromachined structures and an efficient image processing method, and demonstrates that carefully engineered microstructures within the sensor hardware can significantly enhance sensitivity while reducing computational load. Unlike traditional designs with tracking markers, our sensor incorporates an interface surface with micromachined trenches, as an example of microstructures, which modulate light transmission and amplify the variation in response to applied force. By capturing variations in brightness, wire width, and cross pattern locations with a camera, the sensor accurately infers the contact location, the magnitude of displacement and applied force with a lightweight convolutional neural network (CNN). Theoretical and experimental results demonstrated that the microstructures significantly enhance sensitivity by amplifying the visual effects of shape distortion. The sensor system effectively detected forces below 10 mN, and achieved a millimetre-level single-point spatial resolution. Using a model with only one convolutional layer, a mean absolute error (MAE) below 0.05 mm have been achieved. Its soft sensor body ensures compatibility with soft robots and wearable electronics, while its immunity to electrical crosstalk and interference guarantees reliability in complex human-machine environments.
Benchmarking Graph Learning for Drug-Drug Interaction Prediction
Oct 24, 2024Abstract:Predicting drug-drug interaction (DDI) plays an important role in pharmacology and healthcare for identifying potential adverse interactions and beneficial combination therapies between drug pairs. Recently, a flurry of graph learning methods have been introduced to predict drug-drug interactions. However, evaluating existing methods has several limitations, such as the absence of a unified comparison framework for DDI prediction methods, lack of assessments in meaningful real-world scenarios, and insufficient exploration of side information usage. In order to address these unresolved limitations in the literature, we propose a DDI prediction benchmark on graph learning. We first conduct unified evaluation comparison among existing methods. To meet realistic scenarios, we further evaluate the performance of different methods in settings with new drugs involved and examine the performance across different DDI types. Component analysis is conducted on the biomedical network to better utilize side information. Through this work, we hope to provide more insights for the problem of DDI prediction. Our implementation and data is open-sourced at https://anonymous.4open.science/r/DDI-Benchmark-ACD9/.
Knowledge-Aware Parsimony Learning: A Perspective from Relational Graphs
Jun 29, 2024Abstract:The scaling law, a strategy that involves the brute-force scaling of the training dataset and learnable parameters, has become a prevalent approach for developing stronger learning models. In this paper, we examine its rationale in terms of learning from relational graphs. We demonstrate that directly adhering to such a scaling law does not necessarily yield stronger models due to architectural incompatibility and representation bottlenecks. To tackle this challenge, we propose a novel framework for learning from relational graphs via knowledge-aware parsimony learning. Our method draws inspiration from the duality between data and knowledge inherent in these graphs. Specifically, we first extract knowledge (like symbolic logic and physical laws) during the learning process, and then apply combinatorial generalization to the task at hand. This extracted knowledge serves as the ``building blocks'' for achieving parsimony learning. By applying this philosophy to architecture, parameters, and inference, we can effectively achieve versatile, sample-efficient, and interpretable learning. Experimental results show that our proposed framework surpasses methods that strictly follow the traditional scaling-up roadmap. This highlights the importance of incorporating knowledge in the development of next-generation learning technologies.
R^2AG: Incorporating Retrieval Information into Retrieval Augmented Generation
Jun 19, 2024
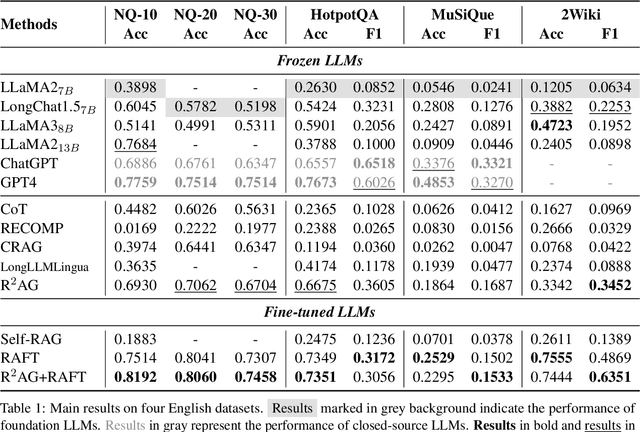

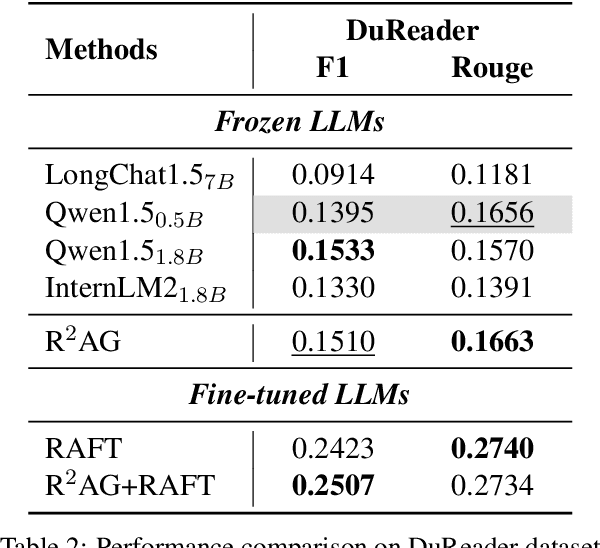
Abstract:Retrieval augmented generation (RAG) has been applied in many scenarios to augment large language models (LLMs) with external documents provided by retrievers. However, a semantic gap exists between LLMs and retrievers due to differences in their training objectives and architectures. This misalignment forces LLMs to passively accept the documents provided by the retrievers, leading to incomprehension in the generation process, where the LLMs are burdened with the task of distinguishing these documents using their inherent knowledge. This paper proposes R$^2$AG, a novel enhanced RAG framework to fill this gap by incorporating Retrieval information into Retrieval Augmented Generation. Specifically, R$^2$AG utilizes the nuanced features from the retrievers and employs a R$^2$-Former to capture retrieval information. Then, a retrieval-aware prompting strategy is designed to integrate retrieval information into LLMs' generation. Notably, R$^2$AG suits low-source scenarios where LLMs and retrievers are frozen. Extensive experiments across five datasets validate the effectiveness, robustness, and efficiency of R$^2$AG. Our analysis reveals that retrieval information serves as an anchor to aid LLMs in the generation process, thereby filling the semantic gap.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge