Ye Wu
PubSub-VFL: Towards Efficient Two-Party Split Learning in Heterogeneous Environments via Publisher/Subscriber Architecture
Oct 14, 2025Abstract:With the rapid advancement of the digital economy, data collaboration between organizations has become a well-established business model, driving the growth of various industries. However, privacy concerns make direct data sharing impractical. To address this, Two-Party Split Learning (a.k.a. Vertical Federated Learning (VFL)) has emerged as a promising solution for secure collaborative learning. Despite its advantages, this architecture still suffers from low computational resource utilization and training efficiency. Specifically, its synchronous dependency design increases training latency, while resource and data heterogeneity among participants further hinder efficient computation. To overcome these challenges, we propose PubSub-VFL, a novel VFL paradigm with a Publisher/Subscriber architecture optimized for two-party collaborative learning with high computational efficiency. PubSub-VFL leverages the decoupling capabilities of the Pub/Sub architecture and the data parallelism of the parameter server architecture to design a hierarchical asynchronous mechanism, reducing training latency and improving system efficiency. Additionally, to mitigate the training imbalance caused by resource and data heterogeneity, we formalize an optimization problem based on participants' system profiles, enabling the selection of optimal hyperparameters while preserving privacy. We conduct a theoretical analysis to demonstrate that PubSub-VFL achieves stable convergence and is compatible with security protocols such as differential privacy. Extensive case studies on five benchmark datasets further validate its effectiveness, showing that, compared to state-of-the-art baselines, PubSub-VFL not only accelerates training by $2 \sim 7\times$ without compromising accuracy, but also achieves a computational resource utilization rate of up to 91.07%.
DDTracking: A Deep Generative Framework for Diffusion MRI Tractography with Streamline Local-Global Spatiotemporal Modeling
Aug 06, 2025Abstract:This paper presents DDTracking, a novel deep generative framework for diffusion MRI tractography that formulates streamline propagation as a conditional denoising diffusion process. In DDTracking, we introduce a dual-pathway encoding network that jointly models local spatial encoding (capturing fine-scale structural details at each streamline point) and global temporal dependencies (ensuring long-range consistency across the entire streamline). Furthermore, we design a conditional diffusion model module, which leverages the learned local and global embeddings to predict streamline propagation orientations for tractography in an end-to-end trainable manner. We conduct a comprehensive evaluation across diverse, independently acquired dMRI datasets, including both synthetic and clinical data. Experiments on two well-established benchmarks with ground truth (ISMRM Challenge and TractoInferno) demonstrate that DDTracking largely outperforms current state-of-the-art tractography methods. Furthermore, our results highlight DDTracking's strong generalizability across heterogeneous datasets, spanning varying health conditions, age groups, imaging protocols, and scanner types. Collectively, DDTracking offers anatomically plausible and robust tractography, presenting a scalable, adaptable, and end-to-end learnable solution for broad dMRI applications. Code is available at: https://github.com/yishengpoxiao/DDtracking.git
Medical Image De-Identification Benchmark Challenge
Jul 31, 2025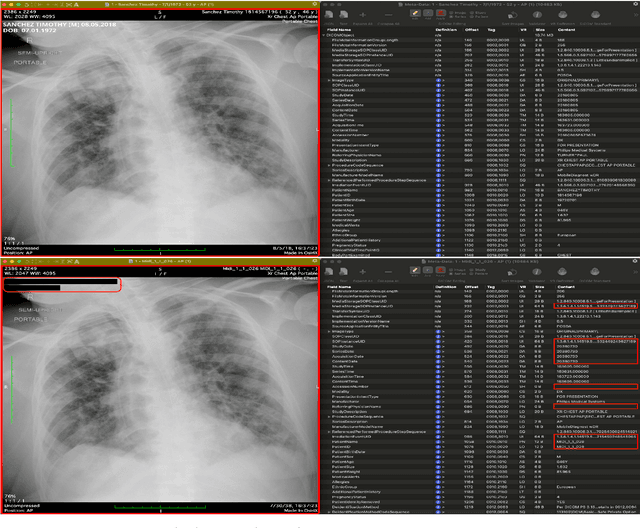
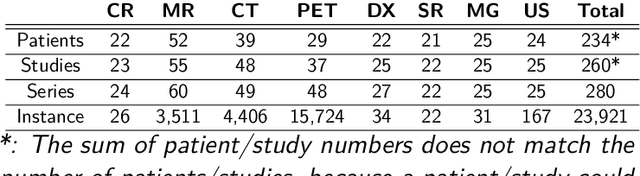
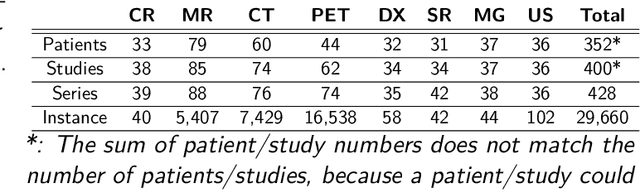
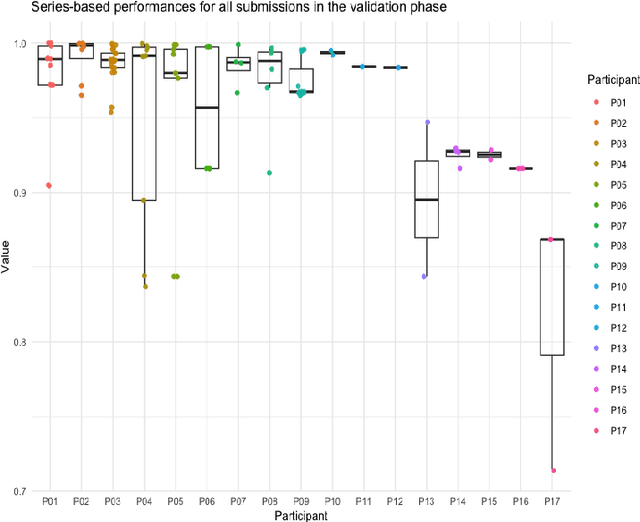
Abstract:The de-identification (deID) of protected health information (PHI) and personally identifiable information (PII) is a fundamental requirement for sharing medical images, particularly through public repositories, to ensure compliance with patient privacy laws. In addition, preservation of non-PHI metadata to inform and enable downstream development of imaging artificial intelligence (AI) is an important consideration in biomedical research. The goal of MIDI-B was to provide a standardized platform for benchmarking of DICOM image deID tools based on a set of rules conformant to the HIPAA Safe Harbor regulation, the DICOM Attribute Confidentiality Profiles, and best practices in preservation of research-critical metadata, as defined by The Cancer Imaging Archive (TCIA). The challenge employed a large, diverse, multi-center, and multi-modality set of real de-identified radiology images with synthetic PHI/PII inserted. The MIDI-B Challenge consisted of three phases: training, validation, and test. Eighty individuals registered for the challenge. In the training phase, we encouraged participants to tune their algorithms using their in-house or public data. The validation and test phases utilized the DICOM images containing synthetic identifiers (of 216 and 322 subjects, respectively). Ten teams successfully completed the test phase of the challenge. To measure success of a rule-based approach to image deID, scores were computed as the percentage of correct actions from the total number of required actions. The scores ranged from 97.91% to 99.93%. Participants employed a variety of open-source and proprietary tools with customized configurations, large language models, and optical character recognition (OCR). In this paper we provide a comprehensive report on the MIDI-B Challenge's design, implementation, results, and lessons learned.
SimWorld: A Unified Benchmark for Simulator-Conditioned Scene Generation via World Model
Mar 18, 2025



Abstract:With the rapid advancement of autonomous driving technology, a lack of data has become a major obstacle to enhancing perception model accuracy. Researchers are now exploring controllable data generation using world models to diversify datasets. However, previous work has been limited to studying image generation quality on specific public datasets. There is still relatively little research on how to build data generation engines for real-world application scenes to achieve large-scale data generation for challenging scenes. In this paper, a simulator-conditioned scene generation engine based on world model is proposed. By constructing a simulation system consistent with real-world scenes, simulation data and labels, which serve as the conditions for data generation in the world model, for any scenes can be collected. It is a novel data generation pipeline by combining the powerful scene simulation capabilities of the simulation engine with the robust data generation capabilities of the world model. In addition, a benchmark with proportionally constructed virtual and real data, is provided for exploring the capabilities of world models in real-world scenes. Quantitative results show that these generated images significantly improve downstream perception models performance. Finally, we explored the generative performance of the world model in urban autonomous driving scenarios. All the data and code will be available at https://github.com/Li-Zn-H/SimWorld.
Deep Learning-Based Diffusion MRI Tractography: Integrating Spatial and Anatomical Information
Mar 05, 2025Abstract:Diffusion MRI tractography technique enables non-invasive visualization of the white matter pathways in the brain. It plays a crucial role in neuroscience and clinical fields by facilitating the study of brain connectivity and neurological disorders. However, the accuracy of reconstructed tractograms has been a longstanding challenge. Recently, deep learning methods have been applied to improve tractograms for better white matter coverage, but often comes at the expense of generating excessive false-positive connections. This is largely due to their reliance on local information to predict long range streamlines. To improve the accuracy of streamline propagation predictions, we introduce a novel deep learning framework that integrates image-domain spatial information and anatomical information along tracts, with the former extracted through convolutional layers and the later modeled via a Transformer-decoder. Additionally, we employ a weighted loss function to address fiber class imbalance encountered during training. We evaluate the proposed method on the simulated ISMRM 2015 Tractography Challenge dataset, achieving a valid streamline rate of 66.2%, white matter coverage of 63.8%, and successfully reconstructing 24 out of 25 bundles. Furthermore, on the multi-site Tractoinferno dataset, the proposed method demonstrates its ability to handle various diffusion MRI acquisition schemes, achieving a 5.7% increase in white matter coverage and a 4.1% decrease in overreach compared to RNN-based methods.
A Novel Streamline-based diffusion MRI Tractography Registration Method with Probabilistic Keypoint Detection
Mar 04, 2025


Abstract:Registration of diffusion MRI tractography is an essential step for analyzing group similarities and variations in the brain's white matter (WM). Streamline-based registration approaches can leverage the 3D geometric information of fiber pathways to enable spatial alignment after registration. Existing methods usually rely on the optimization of the spatial distances to identify the optimal transformation. However, such methods overlook point connectivity patterns within the streamline itself, limiting their ability to identify anatomical correspondences across tractography datasets. In this work, we propose a novel unsupervised approach using deep learning to perform streamline-based dMRI tractography registration. The overall idea is to identify corresponding keypoint pairs across subjects for spatial alignment of tractography datasets. We model tractography as point clouds to leverage the graph connectivity along streamlines. We propose a novel keypoint detection method for streamlines, framed as a probabilistic classification task to identify anatomically consistent correspondences across unstructured streamline sets. In the experiments, we compare several existing methods and show highly effective and efficient tractography registration performance.
What are Models Thinking about? Understanding Large Language Model Hallucinations "Psychology" through Model Inner State Analysis
Feb 19, 2025Abstract:Large language model (LLM) systems suffer from the models' unstable ability to generate valid and factual content, resulting in hallucination generation. Current hallucination detection methods heavily rely on out-of-model information sources, such as RAG to assist the detection, thus bringing heavy additional latency. Recently, internal states of LLMs' inference have been widely used in numerous research works, such as prompt injection detection, etc. Considering the interpretability of LLM internal states and the fact that they do not require external information sources, we introduce such states into LLM hallucination detection. In this paper, we systematically analyze different internal states' revealing features during inference forward and comprehensively evaluate their ability in hallucination detection. Specifically, we cut the forward process of a large language model into three stages: understanding, query, generation, and extracting the internal state from these stages. By analyzing these states, we provide a deep understanding of why the hallucinated content is generated and what happened in the internal state of the models. Then, we introduce these internal states into hallucination detection and conduct comprehensive experiments to discuss the advantages and limitations.
MICCAI-CDMRI 2023 QuantConn Challenge Findings on Achieving Robust Quantitative Connectivity through Harmonized Preprocessing of Diffusion MRI
Nov 14, 2024



Abstract:White matter alterations are increasingly implicated in neurological diseases and their progression. International-scale studies use diffusion-weighted magnetic resonance imaging (DW-MRI) to qualitatively identify changes in white matter microstructure and connectivity. Yet, quantitative analysis of DW-MRI data is hindered by inconsistencies stemming from varying acquisition protocols. There is a pressing need to harmonize the preprocessing of DW-MRI datasets to ensure the derivation of robust quantitative diffusion metrics across acquisitions. In the MICCAI-CDMRI 2023 QuantConn challenge, participants were provided raw data from the same individuals collected on the same scanner but with two different acquisitions and tasked with preprocessing the DW-MRI to minimize acquisition differences while retaining biological variation. Submissions are evaluated on the reproducibility and comparability of cross-acquisition bundle-wise microstructure measures, bundle shape features, and connectomics. The key innovations of the QuantConn challenge are that (1) we assess bundles and tractography in the context of harmonization for the first time, (2) we assess connectomics in the context of harmonization for the first time, and (3) we have 10x additional subjects over prior harmonization challenge, MUSHAC and 100x over SuperMUDI. We find that bundle surface area, fractional anisotropy, connectome assortativity, betweenness centrality, edge count, modularity, nodal strength, and participation coefficient measures are most biased by acquisition and that machine learning voxel-wise correction, RISH mapping, and NeSH methods effectively reduce these biases. In addition, microstructure measures AD, MD, RD, bundle length, connectome density, efficiency, and path length are least biased by these acquisition differences.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024/019
EVENet: Evidence-based Ensemble Learning for Uncertainty-aware Brain Parcellation Using Diffusion MRI
Sep 11, 2024



Abstract:In this study, we developed an Evidence-based Ensemble Neural Network, namely EVENet, for anatomical brain parcellation using diffusion MRI. The key innovation of EVENet is the design of an evidential deep learning framework to quantify predictive uncertainty at each voxel during a single inference. Using EVENet, we obtained accurate parcellation and uncertainty estimates across different datasets from healthy and clinical populations and with different imaging acquisitions. The overall network includes five parallel subnetworks, where each is dedicated to learning the FreeSurfer parcellation for a certain diffusion MRI parameter. An evidence-based ensemble methodology is then proposed to fuse the individual outputs. We perform experimental evaluations on large-scale datasets from multiple imaging sources, including high-quality diffusion MRI data from healthy adults and clinically diffusion MRI data from participants with various brain diseases (schizophrenia, bipolar disorder, attention-deficit/hyperactivity disorder, Parkinson's disease, cerebral small vessel disease, and neurosurgical patients with brain tumors). Compared to several state-of-the-art methods, our experimental results demonstrate highly improved parcellation accuracy across the multiple testing datasets despite the differences in dMRI acquisition protocols and health conditions. Furthermore, thanks to the uncertainty estimation, our EVENet approach demonstrates a good ability to detect abnormal brain regions in patients with lesions, enhancing the interpretability and reliability of the segmentation results.
Reliable Deep Diffusion Tensor Estimation: Rethinking the Power of Data-Driven Optimization Routine
Sep 04, 2024



Abstract:Diffusion tensor imaging (DTI) holds significant importance in clinical diagnosis and neuroscience research. However, conventional model-based fitting methods often suffer from sensitivity to noise, leading to decreased accuracy in estimating DTI parameters. While traditional data-driven deep learning methods have shown potential in terms of accuracy and efficiency, their limited generalization to out-of-training-distribution data impedes their broader application due to the diverse scan protocols used across centers, scanners, and studies. This work aims to tackle these challenges and promote the use of DTI by introducing a data-driven optimization-based method termed DoDTI. DoDTI combines the weighted linear least squares fitting algorithm and regularization by denoising technique. The former fits DW images from diverse acquisition settings into diffusion tensor field, while the latter applies a deep learning-based denoiser to regularize the diffusion tensor field instead of the DW images, which is free from the limitation of fixed-channel assignment of the network. The optimization object is solved using the alternating direction method of multipliers and then unrolled to construct a deep neural network, leveraging a data-driven strategy to learn network parameters. Extensive validation experiments are conducted utilizing both internally simulated datasets and externally obtained in-vivo datasets. The results, encompassing both qualitative and quantitative analyses, showcase that the proposed method attains state-of-the-art performance in DTI parameter estimation. Notably, it demonstrates superior generalization, accuracy, and efficiency, rendering it highly reliable for widespread application in the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge