Yunqi Gu
AllTracker: Efficient Dense Point Tracking at High Resolution
Jun 08, 2025Abstract:We introduce AllTracker: a model that estimates long-range point tracks by way of estimating the flow field between a query frame and every other frame of a video. Unlike existing point tracking methods, our approach delivers high-resolution and dense (all-pixel) correspondence fields, which can be visualized as flow maps. Unlike existing optical flow methods, our approach corresponds one frame to hundreds of subsequent frames, rather than just the next frame. We develop a new architecture for this task, blending techniques from existing work in optical flow and point tracking: the model performs iterative inference on low-resolution grids of correspondence estimates, propagating information spatially via 2D convolution layers, and propagating information temporally via pixel-aligned attention layers. The model is fast and parameter-efficient (16 million parameters), and delivers state-of-the-art point tracking accuracy at high resolution (i.e., tracking 768x1024 pixels, on a 40G GPU). A benefit of our design is that we can train on a wider set of datasets, and we find that doing so is crucial for top performance. We provide an extensive ablation study on our architecture details and training recipe, making it clear which details matter most. Our code and model weights are available at https://alltracker.github.io .
BlenderGym: Benchmarking Foundational Model Systems for Graphics Editing
Apr 02, 2025



Abstract:3D graphics editing is crucial in applications like movie production and game design, yet it remains a time-consuming process that demands highly specialized domain expertise. Automating this process is challenging because graphical editing requires performing a variety of tasks, each requiring distinct skill sets. Recently, vision-language models (VLMs) have emerged as a powerful framework for automating the editing process, but their development and evaluation are bottlenecked by the lack of a comprehensive benchmark that requires human-level perception and presents real-world editing complexity. In this work, we present BlenderGym, the first comprehensive VLM system benchmark for 3D graphics editing. BlenderGym evaluates VLM systems through code-based 3D reconstruction tasks. We evaluate closed- and open-source VLM systems and observe that even the state-of-the-art VLM system struggles with tasks relatively easy for human Blender users. Enabled by BlenderGym, we study how inference scaling techniques impact VLM's performance on graphics editing tasks. Notably, our findings reveal that the verifier used to guide the scaling of generation can itself be improved through inference scaling, complementing recent insights on inference scaling of LLM generation in coding and math tasks. We further show that inference compute is not uniformly effective and can be optimized by strategically distributing it between generation and verification.
Dual-scale Enhanced and Cross-generative Consistency Learning for Semi-supervised Polyp Segmentation
Dec 26, 2023



Abstract:Automatic polyp segmentation plays a crucial role in the early diagnosis and treatment of colorectal cancer (CRC). However, existing methods heavily rely on fully supervised training, which requires a large amount of labeled data with time-consuming pixel-wise annotations. Moreover, accurately segmenting polyps poses challenges due to variations in shape, size, and location. To address these issues, we propose a novel Dual-scale Enhanced and Cross-generative consistency learning framework for semi-supervised polyp Segmentation (DEC-Seg) from colonoscopy images. First, we propose a Cross-level Feature Aggregation (CFA) module that integrates cross-level adjacent layers to enhance the feature representation ability across different resolutions. To address scale variation, we present a scale-enhanced consistency constraint, which ensures consistency in the segmentation maps generated from the same input image at different scales. This constraint helps handle variations in polyp sizes and improves the robustness of the model. Additionally, we design a scale-aware perturbation consistency scheme to enhance the robustness of the mean teacher model. Furthermore, we propose a cross-generative consistency scheme, in which the original and perturbed images can be reconstructed using cross-segmentation maps. This consistency constraint allows us to mine effective feature representations and boost the segmentation performance. To produce more accurate segmentation maps, we propose a Dual-scale Complementary Fusion (DCF) module that integrates features from two scale-specific decoders operating at different scales. Extensive experimental results on five benchmark datasets demonstrate the effectiveness of our DEC-Seg against other state-of-the-art semi-supervised segmentation approaches. The implementation code will be released at https://github.com/taozh2017/DECSeg.
SegRap2023: A Benchmark of Organs-at-Risk and Gross Tumor Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Dec 15, 2023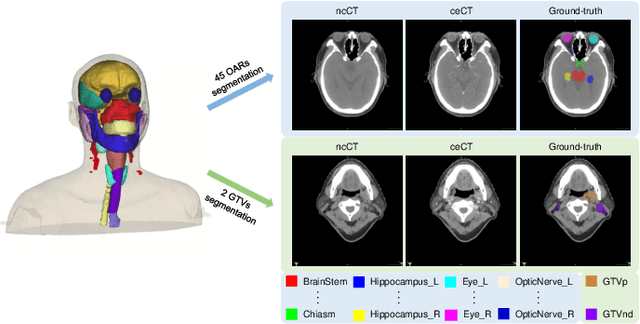
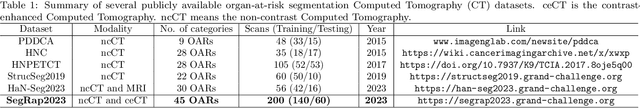
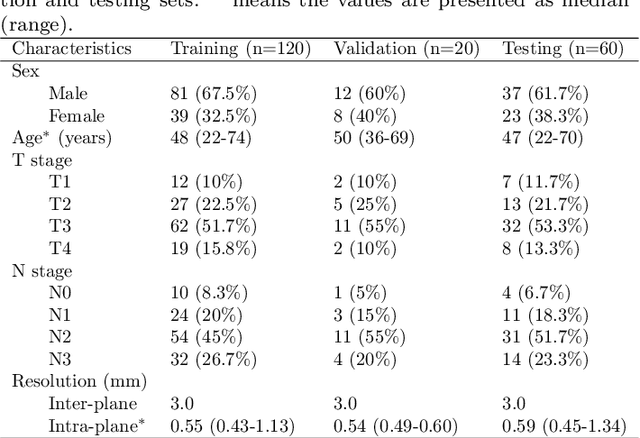
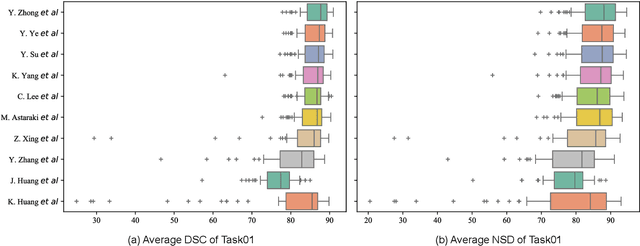
Abstract:Radiation therapy is a primary and effective NasoPharyngeal Carcinoma (NPC) treatment strategy. The precise delineation of Gross Tumor Volumes (GTVs) and Organs-At-Risk (OARs) is crucial in radiation treatment, directly impacting patient prognosis. Previously, the delineation of GTVs and OARs was performed by experienced radiation oncologists. Recently, deep learning has achieved promising results in many medical image segmentation tasks. However, for NPC OARs and GTVs segmentation, few public datasets are available for model development and evaluation. To alleviate this problem, the SegRap2023 challenge was organized in conjunction with MICCAI2023 and presented a large-scale benchmark for OAR and GTV segmentation with 400 Computed Tomography (CT) scans from 200 NPC patients, each with a pair of pre-aligned non-contrast and contrast-enhanced CT scans. The challenge's goal was to segment 45 OARs and 2 GTVs from the paired CT scans. In this paper, we detail the challenge and analyze the solutions of all participants. The average Dice similarity coefficient scores for all submissions ranged from 76.68\% to 86.70\%, and 70.42\% to 73.44\% for OARs and GTVs, respectively. We conclude that the segmentation of large-size OARs is well-addressed, and more efforts are needed for GTVs and small-size or thin-structure OARs. The benchmark will remain publicly available here: https://segrap2023.grand-challenge.org
Segment Anything Model-guided Collaborative Learning Network for Scribble-supervised Polyp Segmentation
Dec 01, 2023Abstract:Polyp segmentation plays a vital role in accurately locating polyps at an early stage, which holds significant clinical importance for the prevention of colorectal cancer. Various polyp segmentation methods have been developed using fully-supervised deep learning techniques. However, pixel-wise annotation for polyp images by physicians during the diagnosis is both time-consuming and expensive. Moreover, visual foundation models such as the Segment Anything Model (SAM) have shown remarkable performance. Nevertheless, directly applying SAM to medical segmentation may not produce satisfactory results due to the inherent absence of medical knowledge. In this paper, we propose a novel SAM-guided Collaborative Learning Network (SAM-CLNet) for scribble-supervised polyp segmentation, enabling a collaborative learning process between our segmentation network and SAM to boost the model performance. Specifically, we first propose a Cross-level Enhancement and Aggregation Network (CEA-Net) for weakly-supervised polyp segmentation. Within CEA-Net, we propose a Cross-level Enhancement Module (CEM) that integrates the adjacent features to enhance the representation capabilities of different resolution features. Additionally, a Feature Aggregation Module (FAM) is employed to capture richer features across multiple levels. Moreover, we present a box-augmentation strategy that combines the segmentation maps generated by CEA-Net with scribble annotations to create more precise prompts. These prompts are then fed into SAM, generating segmentation SAM-guided masks, which can provide additional supervision to train CEA-Net effectively. Furthermore, we present an Image-level Filtering Mechanism to filter out unreliable SAM-guided masks. Extensive experimental results show that our SAM-CLNet outperforms state-of-the-art weakly-supervised segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge