Shuolin Liu
Multi-Class Segmentation of Aortic Branches and Zones in Computed Tomography Angiography: The AortaSeg24 Challenge
Feb 07, 2025



Abstract:Multi-class segmentation of the aorta in computed tomography angiography (CTA) scans is essential for diagnosing and planning complex endovascular treatments for patients with aortic dissections. However, existing methods reduce aortic segmentation to a binary problem, limiting their ability to measure diameters across different branches and zones. Furthermore, no open-source dataset is currently available to support the development of multi-class aortic segmentation methods. To address this gap, we organized the AortaSeg24 MICCAI Challenge, introducing the first dataset of 100 CTA volumes annotated for 23 clinically relevant aortic branches and zones. This dataset was designed to facilitate both model development and validation. The challenge attracted 121 teams worldwide, with participants leveraging state-of-the-art frameworks such as nnU-Net and exploring novel techniques, including cascaded models, data augmentation strategies, and custom loss functions. We evaluated the submitted algorithms using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD), highlighting the approaches adopted by the top five performing teams. This paper presents the challenge design, dataset details, evaluation metrics, and an in-depth analysis of the top-performing algorithms. The annotated dataset, evaluation code, and implementations of the leading methods are publicly available to support further research. All resources can be accessed at https://aortaseg24.grand-challenge.org.
SegRap2023: A Benchmark of Organs-at-Risk and Gross Tumor Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Dec 15, 2023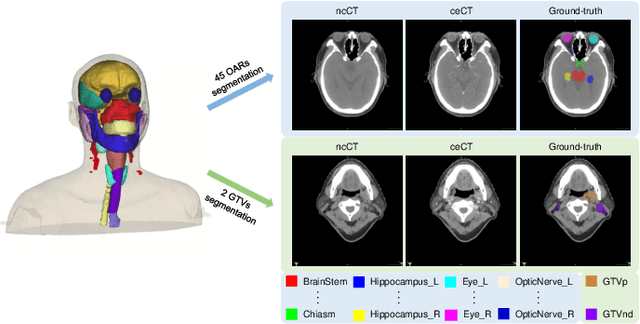
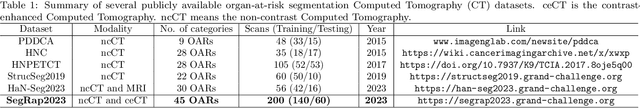
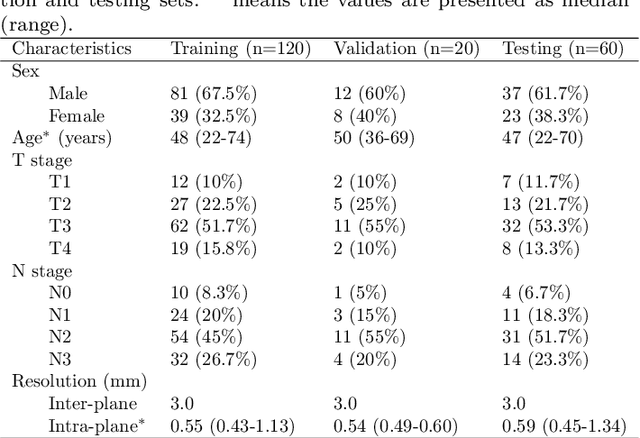
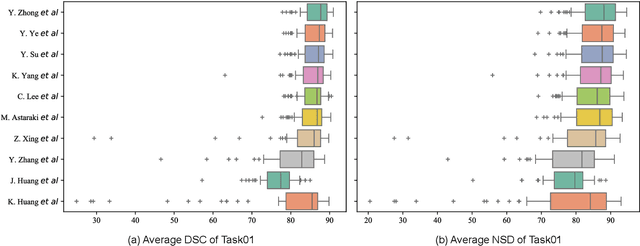
Abstract:Radiation therapy is a primary and effective NasoPharyngeal Carcinoma (NPC) treatment strategy. The precise delineation of Gross Tumor Volumes (GTVs) and Organs-At-Risk (OARs) is crucial in radiation treatment, directly impacting patient prognosis. Previously, the delineation of GTVs and OARs was performed by experienced radiation oncologists. Recently, deep learning has achieved promising results in many medical image segmentation tasks. However, for NPC OARs and GTVs segmentation, few public datasets are available for model development and evaluation. To alleviate this problem, the SegRap2023 challenge was organized in conjunction with MICCAI2023 and presented a large-scale benchmark for OAR and GTV segmentation with 400 Computed Tomography (CT) scans from 200 NPC patients, each with a pair of pre-aligned non-contrast and contrast-enhanced CT scans. The challenge's goal was to segment 45 OARs and 2 GTVs from the paired CT scans. In this paper, we detail the challenge and analyze the solutions of all participants. The average Dice similarity coefficient scores for all submissions ranged from 76.68\% to 86.70\%, and 70.42\% to 73.44\% for OARs and GTVs, respectively. We conclude that the segmentation of large-size OARs is well-addressed, and more efforts are needed for GTVs and small-size or thin-structure OARs. The benchmark will remain publicly available here: https://segrap2023.grand-challenge.org
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
OpenKBP-Opt: An international and reproducible evaluation of 76 knowledge-based planning pipelines
Feb 16, 2022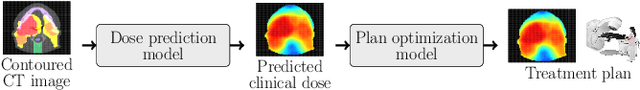

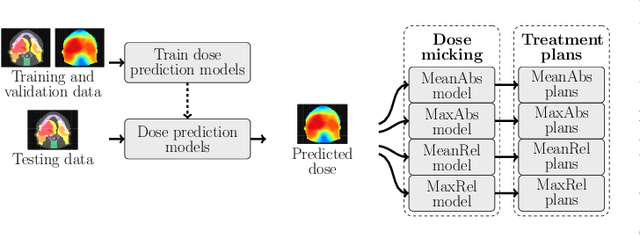
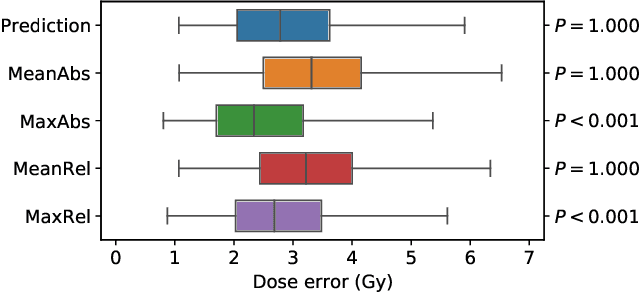
Abstract:We establish an open framework for developing plan optimization models for knowledge-based planning (KBP) in radiotherapy. Our framework includes reference plans for 100 patients with head-and-neck cancer and high-quality dose predictions from 19 KBP models that were developed by different research groups during the OpenKBP Grand Challenge. The dose predictions were input to four optimization models to form 76 unique KBP pipelines that generated 7600 plans. The predictions and plans were compared to the reference plans via: dose score, which is the average mean absolute voxel-by-voxel difference in dose a model achieved; the deviation in dose-volume histogram (DVH) criterion; and the frequency of clinical planning criteria satisfaction. We also performed a theoretical investigation to justify our dose mimicking models. The range in rank order correlation of the dose score between predictions and their KBP pipelines was 0.50 to 0.62, which indicates that the quality of the predictions is generally positively correlated with the quality of the plans. Additionally, compared to the input predictions, the KBP-generated plans performed significantly better (P<0.05; one-sided Wilcoxon test) on 18 of 23 DVH criteria. Similarly, each optimization model generated plans that satisfied a higher percentage of criteria than the reference plans. Lastly, our theoretical investigation demonstrated that the dose mimicking models generated plans that are also optimal for a conventional planning model. This was the largest international effort to date for evaluating the combination of KBP prediction and optimization models. In the interest of reproducibility, our data and code is freely available at https://github.com/ababier/open-kbp-opt.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge