Yiwen Ye
SegRap2025: A Benchmark of Gross Tumor Volume and Lymph Node Clinical Target Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Jan 28, 2026Abstract:Accurate delineation of Gross Tumor Volume (GTV), Lymph Node Clinical Target Volume (LN CTV), and Organ-at-Risk (OAR) from Computed Tomography (CT) scans is essential for precise radiotherapy planning in Nasopharyngeal Carcinoma (NPC). Building upon SegRap2023, which focused on OAR and GTV segmentation using single-center paired non-contrast CT (ncCT) and contrast-enhanced CT (ceCT) scans, the SegRap2025 challenge aims to enhance the generalizability and robustness of segmentation models across imaging centers and modalities. SegRap2025 comprises two tasks: Task01 addresses GTV segmentation using paired CT from the SegRap2023 dataset, with an additional external testing set to evaluate cross-center generalization, and Task02 focuses on LN CTV segmentation using multi-center training data and an unseen external testing set, where each case contains paired CT scans or a single modality, emphasizing both cross-center and cross-modality robustness. This paper presents the challenge setup and provides a comprehensive analysis of the solutions submitted by ten participating teams. For GTV segmentation task, the top-performing models achieved average Dice Similarity Coefficient (DSC) of 74.61% and 56.79% on the internal and external testing cohorts, respectively. For LN CTV segmentation task, the highest average DSC values reached 60.24%, 60.50%, and 57.23% on paired CT, ceCT-only, and ncCT-only subsets, respectively. SegRap2025 establishes a large-scale multi-center, multi-modality benchmark for evaluating the generalization and robustness in radiotherapy target segmentation, providing valuable insights toward clinically applicable automated radiotherapy planning systems. The benchmark is available at: https://hilab-git.github.io/SegRap2025_Challenge.
Unified Start, Personalized End: Progressive Pruning for Efficient 3D Medical Image Segmentation
Sep 11, 2025Abstract:3D medical image segmentation often faces heavy resource and time consumption, limiting its scalability and rapid deployment in clinical environments. Existing efficient segmentation models are typically static and manually designed prior to training, which restricts their adaptability across diverse tasks and makes it difficult to balance performance with resource efficiency. In this paper, we propose PSP-Seg, a progressive pruning framework that enables dynamic and efficient 3D segmentation. PSP-Seg begins with a redundant model and iteratively prunes redundant modules through a combination of block-wise pruning and a functional decoupling loss. We evaluate PSP-Seg on five public datasets, benchmarking it against seven state-of-the-art models and six efficient segmentation models. Results demonstrate that the lightweight variant, PSP-Seg-S, achieves performance on par with nnU-Net while reducing GPU memory usage by 42-45%, training time by 29-48%, and parameter number by 83-87% across all datasets. These findings underscore PSP-Seg's potential as a cost-effective yet high-performing alternative for widespread clinical application.
Enjoying Information Dividend: Gaze Track-based Medical Weakly Supervised Segmentation
May 28, 2025Abstract:Weakly supervised semantic segmentation (WSSS) in medical imaging struggles with effectively using sparse annotations. One promising direction for WSSS leverages gaze annotations, captured via eye trackers that record regions of interest during diagnostic procedures. However, existing gaze-based methods, such as GazeMedSeg, do not fully exploit the rich information embedded in gaze data. In this paper, we propose GradTrack, a framework that utilizes physicians' gaze track, including fixation points, durations, and temporal order, to enhance WSSS performance. GradTrack comprises two key components: Gaze Track Map Generation and Track Attention, which collaboratively enable progressive feature refinement through multi-level gaze supervision during the decoding process. Experiments on the Kvasir-SEG and NCI-ISBI datasets demonstrate that GradTrack consistently outperforms existing gaze-based methods, achieving Dice score improvements of 3.21\% and 2.61\%, respectively. Moreover, GradTrack significantly narrows the performance gap with fully supervised models such as nnUNet.
Meta Curvature-Aware Minimization for Domain Generalization
Dec 16, 2024Abstract:Domain generalization (DG) aims to enhance the ability of models trained on source domains to generalize effectively to unseen domains. Recently, Sharpness-Aware Minimization (SAM) has shown promise in this area by reducing the sharpness of the loss landscape to obtain more generalized models. However, SAM and its variants sometimes fail to guide the model toward a flat minimum, and their training processes exhibit limitations, hindering further improvements in model generalization. In this paper, we first propose an improved model training process aimed at encouraging the model to converge to a flat minima. To achieve this, we design a curvature metric that has a minimal effect when the model is far from convergence but becomes increasingly influential in indicating the curvature of the minima as the model approaches a local minimum. Then we derive a novel algorithm from this metric, called Meta Curvature-Aware Minimization (MeCAM), to minimize the curvature around the local minima. Specifically, the optimization objective of MeCAM simultaneously minimizes the regular training loss, the surrogate gap of SAM, and the surrogate gap of meta-learning. We provide theoretical analysis on MeCAM's generalization error and convergence rate, and demonstrate its superiority over existing DG methods through extensive experiments on five benchmark DG datasets, including PACS, VLCS, OfficeHome, TerraIncognita, and DomainNet. Code will be available on GitHub.
CoSAM: Self-Correcting SAM for Domain Generalization in 2D Medical Image Segmentation
Nov 15, 2024



Abstract:Medical images often exhibit distribution shifts due to variations in imaging protocols and scanners across different medical centers. Domain Generalization (DG) methods aim to train models on source domains that can generalize to unseen target domains. Recently, the segment anything model (SAM) has demonstrated strong generalization capabilities due to its prompt-based design, and has gained significant attention in image segmentation tasks. Existing SAM-based approaches attempt to address the need for manual prompts by introducing prompt generators that automatically generate these prompts. However, we argue that auto-generated prompts may not be sufficiently accurate under distribution shifts, potentially leading to incorrect predictions that still require manual verification and correction by clinicians. To address this challenge, we propose a method for 2D medical image segmentation called Self-Correcting SAM (CoSAM). Our approach begins by generating coarse masks using SAM in a prompt-free manner, providing prior prompts for the subsequent stages, and eliminating the need for prompt generators. To automatically refine these coarse masks, we introduce a generalized error decoder that simulates the correction process typically performed by clinicians. Furthermore, we generate diverse prompts as feedback based on the corrected masks, which are used to iteratively refine the predictions within a self-correcting loop, enhancing the generalization performance of our model. Extensive experiments on two medical image segmentation benchmarks across multiple scenarios demonstrate the superiority of CoSAM over state-of-the-art SAM-based methods.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
Day-Night Adaptation: An Innovative Source-free Adaptation Framework for Medical Image Segmentation
Oct 17, 2024



Abstract:Distribution shifts widely exist in medical images acquired from different medical centers, hindering the deployment of semantic segmentation models trained on data from one center (source domain) to another (target domain). While unsupervised domain adaptation (UDA) has shown significant promise in mitigating these shifts, it poses privacy risks due to sharing data between centers. To facilitate adaptation while preserving data privacy, source-free domain adaptation (SFDA) and test-time adaptation (TTA) have emerged as effective paradigms, relying solely on target domain data. However, the scenarios currently addressed by SFDA and TTA are limited, making them less suitable for clinical applications. In a more realistic clinical scenario, the pre-trained model is deployed in a medical centre to assist with clinical tasks during the day and rest at night. During the daytime process, TTA can be employed to enhance inference performance. During the nighttime process, after collecting the test data from the day, the model can be fine-tuned utilizing SFDA to further adapt to the target domain. With above insights, we propose a novel adaptation framework called Day-Night Adaptation (DyNA). This framework adapts the model to the target domain through day-night loops without requiring access to source data. Specifically, we implement distinct adaptation strategies for daytime and nighttime to better meet the demands of clinical settings. During the daytime, model parameters are frozen, and a specific low-frequency prompt is trained for each test sample. Additionally, we construct a memory bank for prompt initialization and develop a warm-up mechanism to enhance prompt training. During nighttime, we integrate a global student model into the traditional teacher-student self-training paradigm to fine-tune the model while ensuring training stability...
MedUniSeg: 2D and 3D Medical Image Segmentation via a Prompt-driven Universal Model
Oct 08, 2024Abstract:Universal segmentation models offer significant potential in addressing a wide range of tasks by effectively leveraging discrete annotations. As the scope of tasks and modalities expands, it becomes increasingly important to generate and strategically position task- and modal-specific priors within the universal model. However, existing universal models often overlook the correlations between different priors, and the optimal placement and frequency of these priors remain underexplored. In this paper, we introduce MedUniSeg, a prompt-driven universal segmentation model designed for 2D and 3D multi-task segmentation across diverse modalities and domains. MedUniSeg employs multiple modal-specific prompts alongside a universal task prompt to accurately characterize the modalities and tasks. To generate the related priors, we propose the modal map (MMap) and the fusion and selection (FUSE) modules, which transform modal and task prompts into corresponding priors. These modal and task priors are systematically introduced at the start and end of the encoding process. We evaluate MedUniSeg on a comprehensive multi-modal upstream dataset consisting of 17 sub-datasets. The results demonstrate that MedUniSeg achieves superior multi-task segmentation performance, attaining a 1.2% improvement in the mean Dice score across the 17 upstream tasks compared to nnUNet baselines, while using less than 1/10 of the parameters. For tasks that underperform during the initial multi-task joint training, we freeze MedUniSeg and introduce new modules to re-learn these tasks. This approach yields an enhanced version, MedUniSeg*, which consistently outperforms MedUniSeg across all tasks. Moreover, MedUniSeg surpasses advanced self-supervised and supervised pre-trained models on six downstream tasks, establishing itself as a high-quality, highly generalizable pre-trained segmentation model.
Pre-training Everywhere: Parameter-Efficient Fine-Tuning for Medical Image Analysis via Target Parameter Pre-training
Aug 27, 2024Abstract:Parameter-efficient fine-tuning (PEFT) techniques have emerged to address issues of overfitting and high computational costs associated with fully fine-tuning in the paradigm of self-supervised learning. Mainstream methods based on PEFT involve adding a few trainable parameters while keeping the pre-trained parameters of the backbone fixed. These methods achieve comparative, and often superior, performance to fully fine-tuning, demonstrating the powerful representation ability of the pre-trained backbone. Despite its success, these methods typically ignore the initialization of the new parameters, often relying solely on random initialization. We argue that if pre-training is significantly beneficial, it should be applied to all parameters requiring representational capacity. Motivated by this insight, we propose a simple yet effective fine-tuning framework based on Target Parameter Pre-training (TPP). The target parameters refer to the new parameters introduced during fine-tuning. TPP includes an additional stage before PEFT to pre-train these target parameters. During this stage, the pre-trained backbone parameters are frozen, and only the target parameters are trainable. A defined pre-text task is used to encourage the target parameters to learn specific representations of downstream data. When PEFT is subsequently employed, the pre-trained target parameters are loaded to enhance fine-tuning efficiency. The proposed TPP framework is versatile, allowing for the integration of various pretext tasks for pre-training and supporting different PEFT methods as backbones. We evaluated the fine-tining performance of our method using five public datasets, including three modalities and two task types. The results demonstrate that the proposed TPP can be easily integrated into existing PEFT methods, significantly improving performance.
From Few to More: Scribble-based Medical Image Segmentation via Masked Context Modeling and Continuous Pseudo Labels
Aug 23, 2024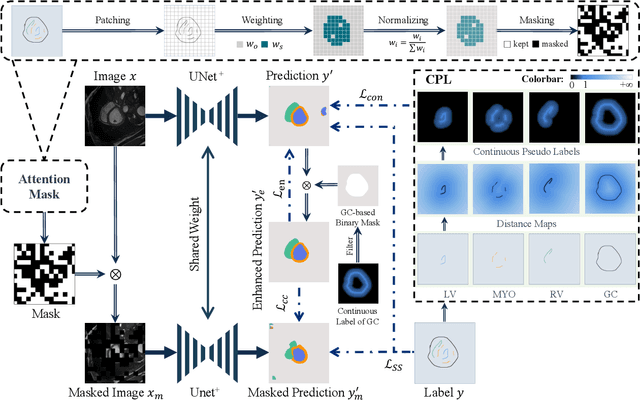
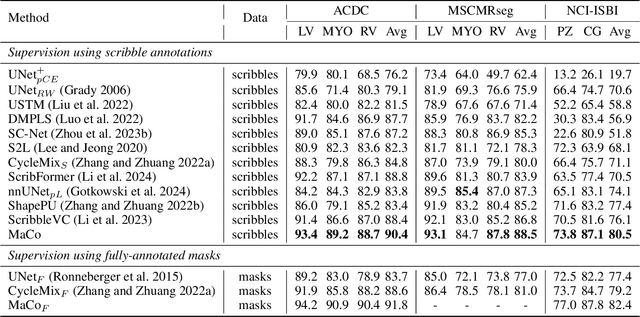
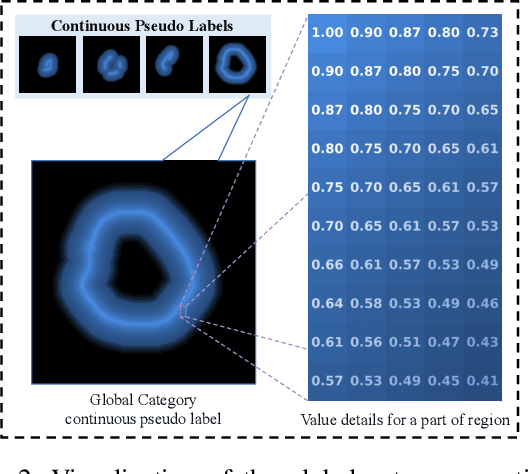
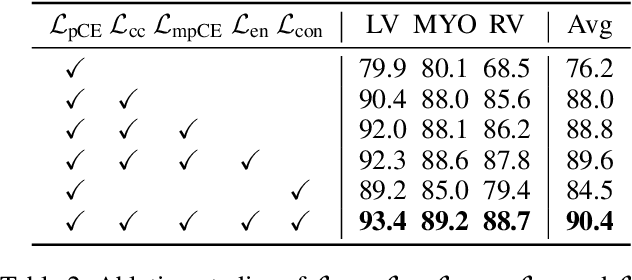
Abstract:Scribble-based weakly supervised segmentation techniques offer comparable performance to fully supervised methods while significantly reducing annotation costs, making them an appealing alternative. Existing methods often rely on auxiliary tasks to enforce semantic consistency and use hard pseudo labels for supervision. However, these methods often overlook the unique requirements of models trained with sparse annotations. Since the model must predict pixel-wise segmentation maps with limited annotations, the ability to handle varying levels of annotation richness is critical. In this paper, we adopt the principle of `from few to more' and propose MaCo, a weakly supervised framework designed for medical image segmentation. MaCo employs masked context modeling (MCM) and continuous pseudo labels (CPL). MCM uses an attention-based masking strategy to disrupt the input image, compelling the model's predictions to remain consistent with those of the original image. CPL converts scribble annotations into continuous pixel-wise labels by applying an exponential decay function to distance maps, resulting in continuous maps that represent the confidence of each pixel belonging to a specific category, rather than using hard pseudo labels. We evaluate MaCo against other weakly supervised methods using three public datasets. The results indicate that MaCo outperforms competing methods across all datasets, setting a new record in weakly supervised medical image segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge