Jianpeng Zhang
UniReg: Foundation Model for Controllable Medical Image Registration
Mar 17, 2025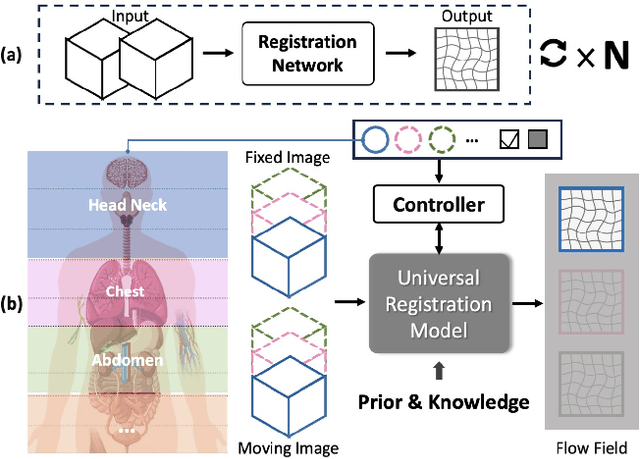
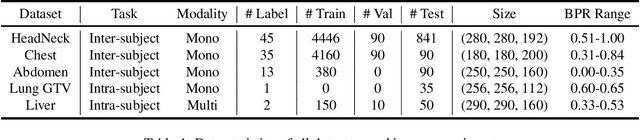
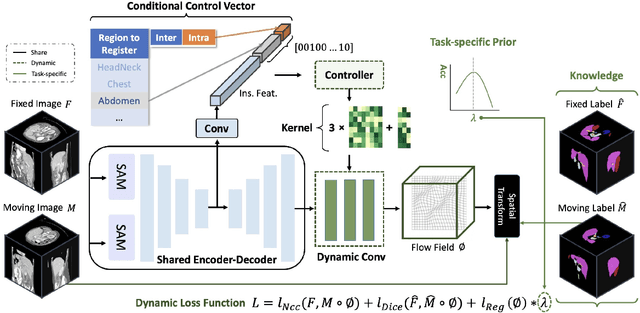
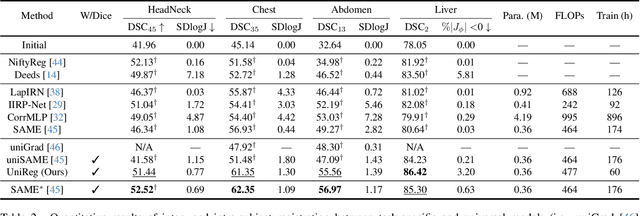
Abstract:Learning-based medical image registration has achieved performance parity with conventional methods while demonstrating a substantial advantage in computational efficiency. However, learning-based registration approaches lack generalizability across diverse clinical scenarios, requiring the laborious development of multiple isolated networks for specific registration tasks, e.g., inter-/intra-subject registration or organ-specific alignment. % To overcome this limitation, we propose \textbf{UniReg}, the first interactive foundation model for medical image registration, which combines the precision advantages of task-specific learning methods with the generalization of traditional optimization methods. Our key innovation is a unified framework for diverse registration scenarios, achieved through a conditional deformation field estimation within a unified registration model. This is realized through a dynamic learning paradigm that explicitly encodes: (1) anatomical structure priors, (2) registration type constraints (inter/intra-subject), and (3) instance-specific features, enabling the generation of scenario-optimal deformation fields. % Through comprehensive experiments encompassing $90$ anatomical structures at different body regions, our UniReg model demonstrates comparable performance with contemporary state-of-the-art methodologies while achieving ~50\% reduction in required training iterations relative to the conventional learning-based paradigm. This optimization contributes to a significant reduction in computational resources, such as training time. Code and model will be available.
Large-scale and Fine-grained Vision-language Pre-training for Enhanced CT Image Understanding
Jan 24, 2025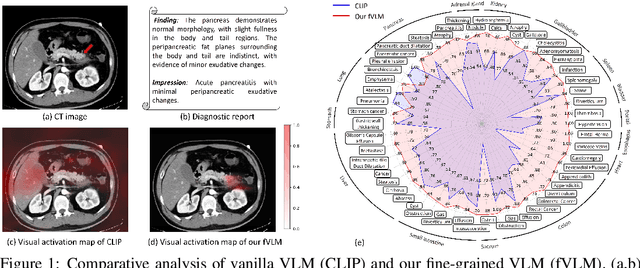
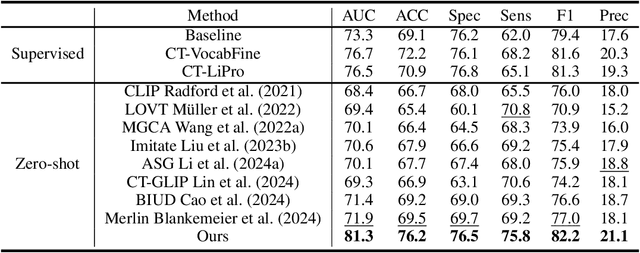
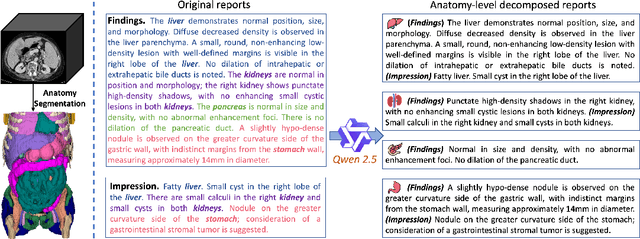
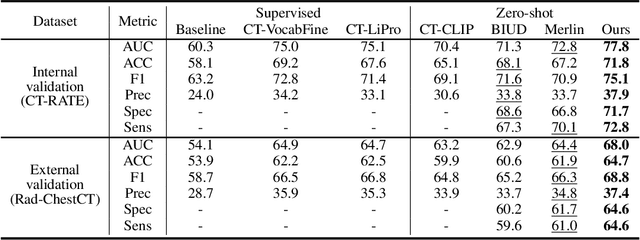
Abstract:Artificial intelligence (AI) shows great potential in assisting radiologists to improve the efficiency and accuracy of medical image interpretation and diagnosis. However, a versatile AI model requires large-scale data and comprehensive annotations, which are often impractical in medical settings. Recent studies leverage radiology reports as a naturally high-quality supervision for medical images, using contrastive language-image pre-training (CLIP) to develop language-informed models for radiological image interpretation. Nonetheless, these approaches typically contrast entire images with reports, neglecting the local associations between imaging regions and report sentences, which may undermine model performance and interoperability. In this paper, we propose a fine-grained vision-language model (fVLM) for anatomy-level CT image interpretation. Specifically, we explicitly match anatomical regions of CT images with corresponding descriptions in radiology reports and perform contrastive pre-training for each anatomy individually. Fine-grained alignment, however, faces considerable false-negative challenges, mainly from the abundance of anatomy-level healthy samples and similarly diseased abnormalities. To tackle this issue, we propose identifying false negatives of both normal and abnormal samples and calibrating contrastive learning from patient-level to disease-aware pairing. We curated the largest CT dataset to date, comprising imaging and report data from 69,086 patients, and conducted a comprehensive evaluation of 54 major and important disease diagnosis tasks across 15 main anatomies. Experimental results demonstrate the substantial potential of fVLM in versatile medical image interpretation. In the zero-shot classification task, we achieved an average AUC of 81.3% on 54 diagnosis tasks, surpassing CLIP and supervised methods by 12.9% and 8.0%, respectively.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
MedUniSeg: 2D and 3D Medical Image Segmentation via a Prompt-driven Universal Model
Oct 08, 2024Abstract:Universal segmentation models offer significant potential in addressing a wide range of tasks by effectively leveraging discrete annotations. As the scope of tasks and modalities expands, it becomes increasingly important to generate and strategically position task- and modal-specific priors within the universal model. However, existing universal models often overlook the correlations between different priors, and the optimal placement and frequency of these priors remain underexplored. In this paper, we introduce MedUniSeg, a prompt-driven universal segmentation model designed for 2D and 3D multi-task segmentation across diverse modalities and domains. MedUniSeg employs multiple modal-specific prompts alongside a universal task prompt to accurately characterize the modalities and tasks. To generate the related priors, we propose the modal map (MMap) and the fusion and selection (FUSE) modules, which transform modal and task prompts into corresponding priors. These modal and task priors are systematically introduced at the start and end of the encoding process. We evaluate MedUniSeg on a comprehensive multi-modal upstream dataset consisting of 17 sub-datasets. The results demonstrate that MedUniSeg achieves superior multi-task segmentation performance, attaining a 1.2% improvement in the mean Dice score across the 17 upstream tasks compared to nnUNet baselines, while using less than 1/10 of the parameters. For tasks that underperform during the initial multi-task joint training, we freeze MedUniSeg and introduce new modules to re-learn these tasks. This approach yields an enhanced version, MedUniSeg*, which consistently outperforms MedUniSeg across all tasks. Moreover, MedUniSeg surpasses advanced self-supervised and supervised pre-trained models on six downstream tasks, establishing itself as a high-quality, highly generalizable pre-trained segmentation model.
Rapid and Accurate Diagnosis of Acute Aortic Syndrome using Non-contrast CT: A Large-scale, Retrospective, Multi-center and AI-based Study
Jun 25, 2024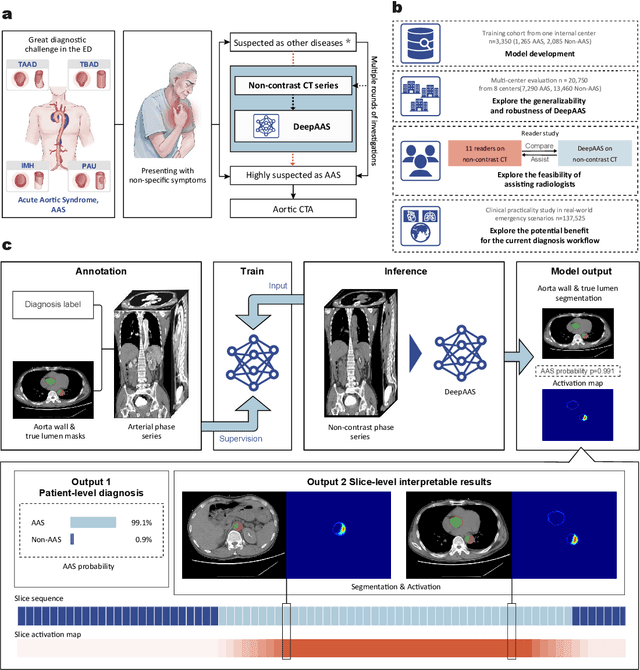
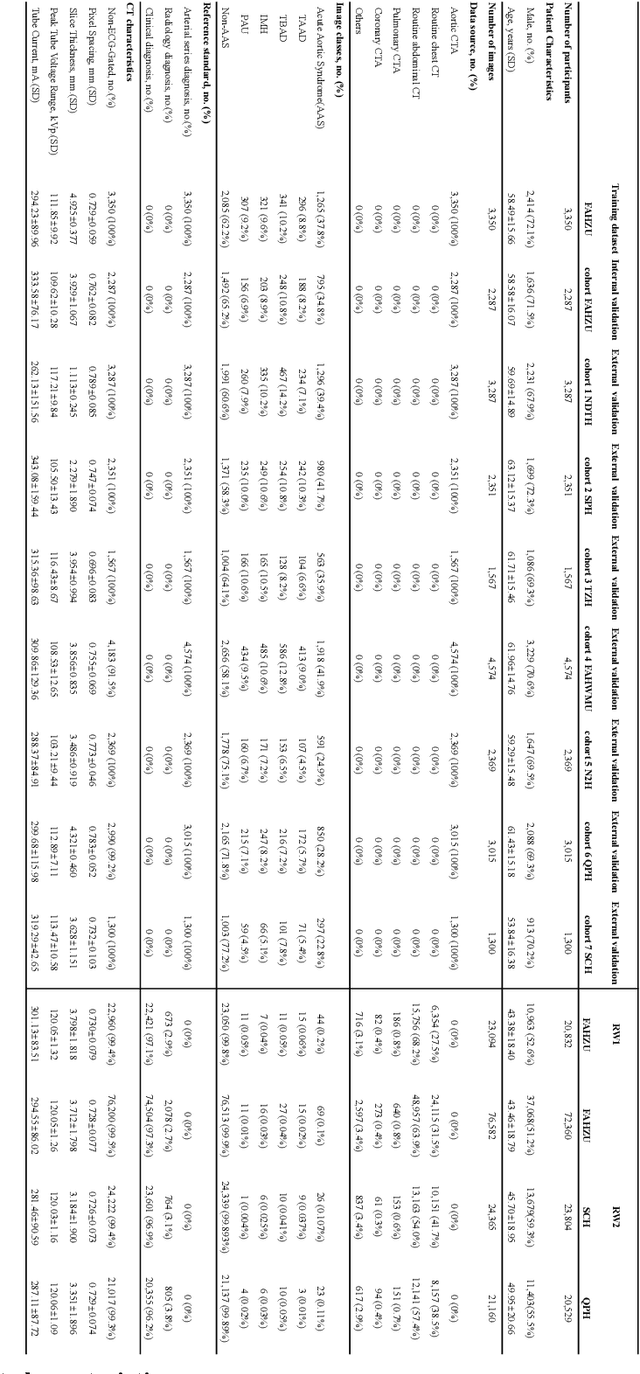
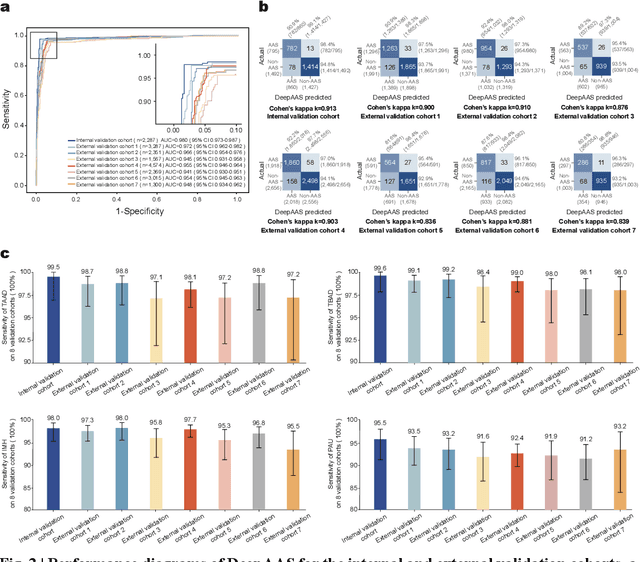
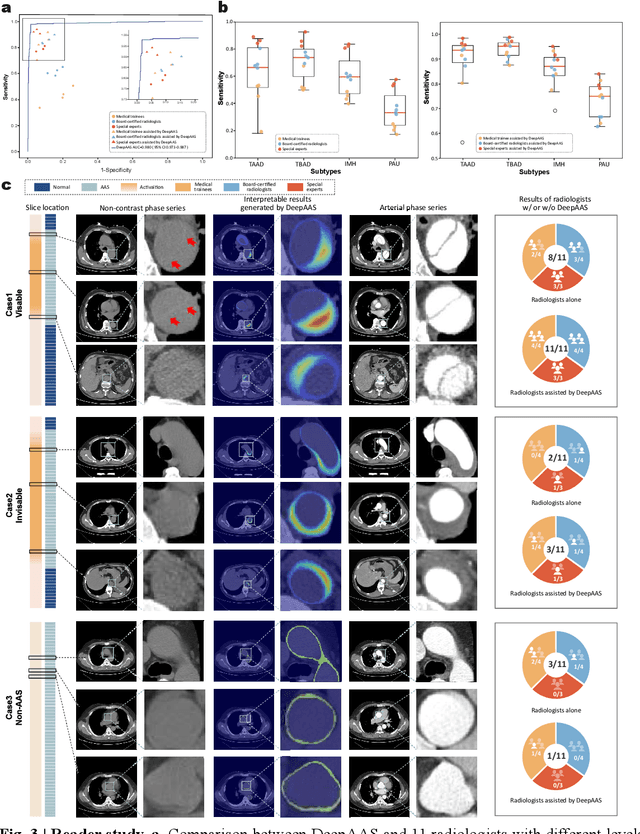
Abstract:Chest pain symptoms are highly prevalent in emergency departments (EDs), where acute aortic syndrome (AAS) is a catastrophic cardiovascular emergency with a high fatality rate, especially when timely and accurate treatment is not administered. However, current triage practices in the ED can cause up to approximately half of patients with AAS to have an initially missed diagnosis or be misdiagnosed as having other acute chest pain conditions. Subsequently, these AAS patients will undergo clinically inaccurate or suboptimal differential diagnosis. Fortunately, even under these suboptimal protocols, nearly all these patients underwent non-contrast CT covering the aorta anatomy at the early stage of differential diagnosis. In this study, we developed an artificial intelligence model (DeepAAS) using non-contrast CT, which is highly accurate for identifying AAS and provides interpretable results to assist in clinical decision-making. Performance was assessed in two major phases: a multi-center retrospective study (n = 20,750) and an exploration in real-world emergency scenarios (n = 137,525). In the multi-center cohort, DeepAAS achieved a mean area under the receiver operating characteristic curve of 0.958 (95% CI 0.950-0.967). In the real-world cohort, DeepAAS detected 109 AAS patients with misguided initial suspicion, achieving 92.6% (95% CI 76.2%-97.5%) in mean sensitivity and 99.2% (95% CI 99.1%-99.3%) in mean specificity. Our AI model performed well on non-contrast CT at all applicable early stages of differential diagnosis workflows, effectively reduced the overall missed diagnosis and misdiagnosis rate from 48.8% to 4.8% and shortened the diagnosis time for patients with misguided initial suspicion from an average of 681.8 (74-11,820) mins to 68.5 (23-195) mins. DeepAAS could effectively fill the gap in the current clinical workflow without requiring additional tests.
CT-GLIP: 3D Grounded Language-Image Pretraining with CT Scans and Radiology Reports for Full-Body Scenarios
Apr 29, 2024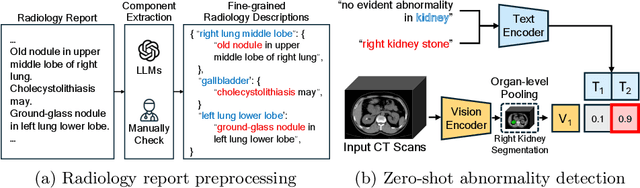
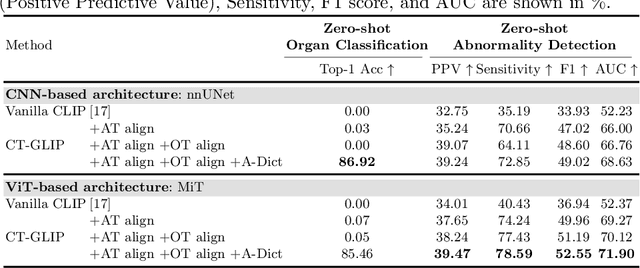
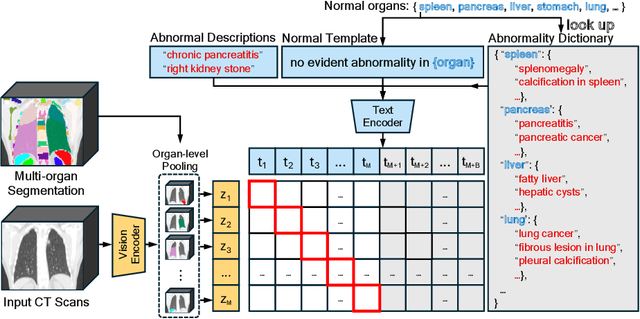
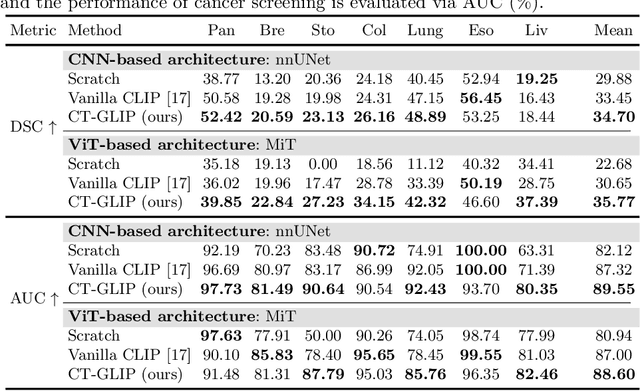
Abstract:Medical Vision-Language Pretraining (Med-VLP) establishes a connection between visual content from medical images and the relevant textual descriptions. Existing Med-VLP methods primarily focus on 2D images depicting a single body part, notably chest X-rays. In this paper, we extend the scope of Med-VLP to encompass 3D images, specifically targeting full-body scenarios, by using a multimodal dataset of CT images and reports. Compared with the 2D counterpart, 3D VLP is required to effectively capture essential semantics from significantly sparser representation in 3D imaging. In this paper, we introduce CT-GLIP (Grounded Language-Image Pretraining with CT scans), a novel method that constructs organ-level image-text pairs to enhance multimodal contrastive learning, aligning grounded visual features with precise diagnostic text. Additionally, we developed an abnormality dictionary to augment contrastive learning with diverse contrastive pairs. Our method, trained on a multimodal CT dataset comprising 44,011 organ-level vision-text pairs from 17,702 patients across 104 organs, demonstrates it can identify organs and abnormalities in a zero-shot manner using natural languages. The performance of CT-GLIP is validated on a separate test set of 1,130 patients, focusing on the 16 most frequent abnormalities across 7 organs. The experimental results show our model's superior performance over the standard CLIP framework across zero-shot and fine-tuning scenarios, using both CNN and ViT architectures.
Bootstrapping Chest CT Image Understanding by Distilling Knowledge from X-ray Expert Models
Apr 07, 2024



Abstract:Radiologists highly desire fully automated versatile AI for medical imaging interpretation. However, the lack of extensively annotated large-scale multi-disease datasets has hindered the achievement of this goal. In this paper, we explore the feasibility of leveraging language as a naturally high-quality supervision for chest CT imaging. In light of the limited availability of image-report pairs, we bootstrap the understanding of 3D chest CT images by distilling chest-related diagnostic knowledge from an extensively pre-trained 2D X-ray expert model. Specifically, we propose a language-guided retrieval method to match each 3D CT image with its semantically closest 2D X-ray image, and perform pair-wise and semantic relation knowledge distillation. Subsequently, we use contrastive learning to align images and reports within the same patient while distinguishing them from the other patients. However, the challenge arises when patients have similar semantic diagnoses, such as healthy patients, potentially confusing if treated as negatives. We introduce a robust contrastive learning that identifies and corrects these false negatives. We train our model with over 12,000 pairs of chest CT images and radiology reports. Extensive experiments across multiple scenarios, including zero-shot learning, report generation, and fine-tuning processes, demonstrate the model's feasibility in interpreting chest CT images.
Modality-Agnostic Structural Image Representation Learning for Deformable Multi-Modality Medical Image Registration
Feb 29, 2024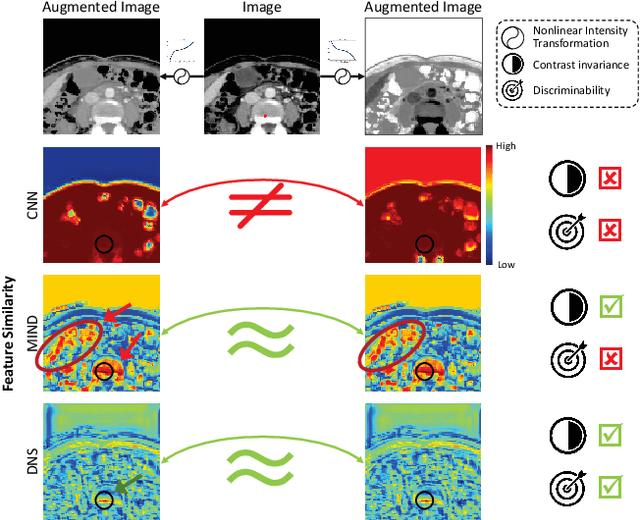
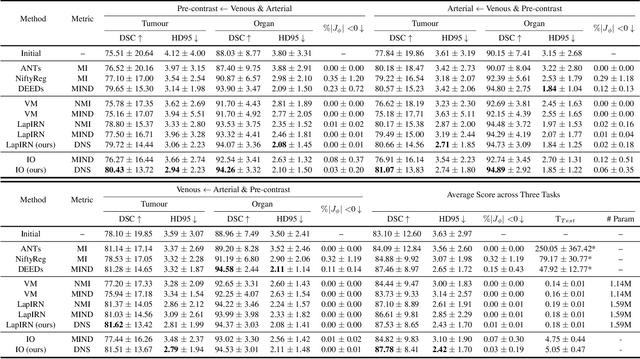
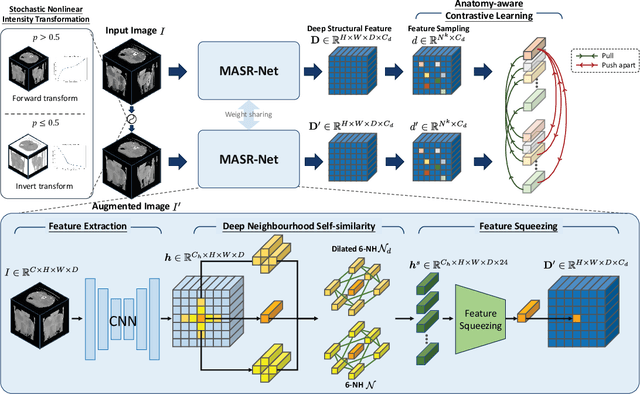
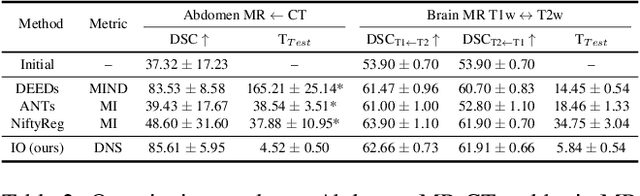
Abstract:Establishing dense anatomical correspondence across distinct imaging modalities is a foundational yet challenging procedure for numerous medical image analysis studies and image-guided radiotherapy. Existing multi-modality image registration algorithms rely on statistical-based similarity measures or local structural image representations. However, the former is sensitive to locally varying noise, while the latter is not discriminative enough to cope with complex anatomical structures in multimodal scans, causing ambiguity in determining the anatomical correspondence across scans with different modalities. In this paper, we propose a modality-agnostic structural representation learning method, which leverages Deep Neighbourhood Self-similarity (DNS) and anatomy-aware contrastive learning to learn discriminative and contrast-invariance deep structural image representations (DSIR) without the need for anatomical delineations or pre-aligned training images. We evaluate our method on multiphase CT, abdomen MR-CT, and brain MR T1w-T2w registration. Comprehensive results demonstrate that our method is superior to the conventional local structural representation and statistical-based similarity measures in terms of discriminability and accuracy.
Continual Self-supervised Learning: Towards Universal Multi-modal Medical Data Representation Learning
Nov 30, 2023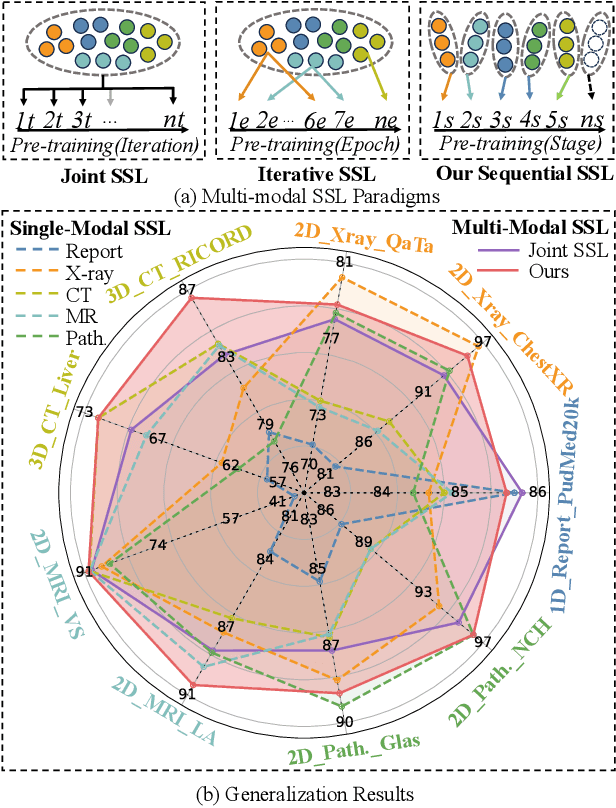
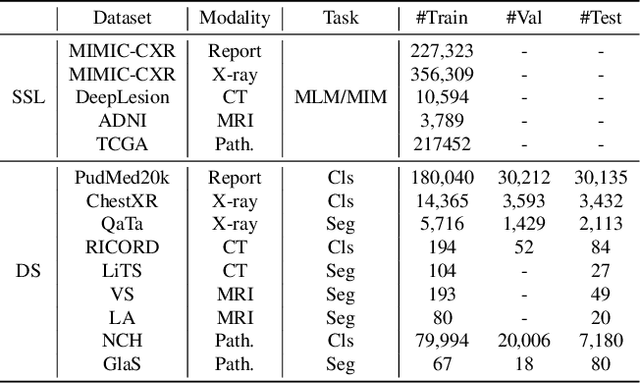
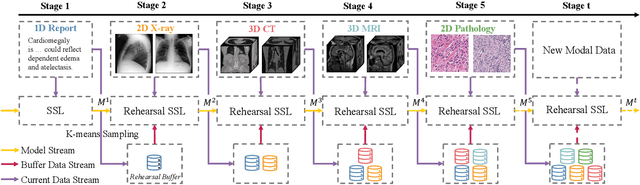
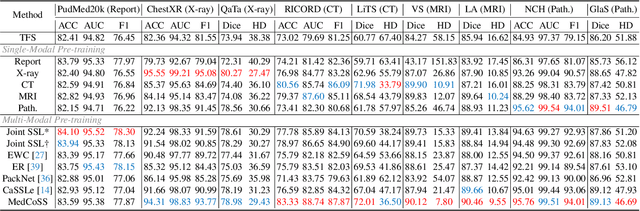
Abstract:Self-supervised learning is an efficient pre-training method for medical image analysis. However, current research is mostly confined to specific-modality data pre-training, consuming considerable time and resources without achieving universality across different modalities. A straightforward solution is combining all modality data for joint self-supervised pre-training, which poses practical challenges. Firstly, our experiments reveal conflicts in representation learning as the number of modalities increases. Secondly, multi-modal data collected in advance cannot cover all real-world scenarios. In this paper, we reconsider versatile self-supervised learning from the perspective of continual learning and propose MedCoSS, a continuous self-supervised learning approach for multi-modal medical data. Unlike joint self-supervised learning, MedCoSS assigns different modality data to different training stages, forming a multi-stage pre-training process. To balance modal conflicts and prevent catastrophic forgetting, we propose a rehearsal-based continual learning method. We introduce the k-means sampling strategy to retain data from previous modalities and rehearse it when learning new modalities. Instead of executing the pretext task on buffer data, a feature distillation strategy and an intra-modal mixup strategy are applied to these data for knowledge retention. We conduct continuous self-supervised pre-training on a large-scale multi-modal unlabeled dataset, including clinical reports, X-rays, CT scans, MRI scans, and pathological images. Experimental results demonstrate MedCoSS's exceptional generalization ability across nine downstream datasets and its significant scalability in integrating new modality data. Code and pre-trained weight are available at https://github.com/yeerwen/MedCoSS.
Parse and Recall: Towards Accurate Lung Nodule Malignancy Prediction like Radiologists
Jul 20, 2023



Abstract:Lung cancer is a leading cause of death worldwide and early screening is critical for improving survival outcomes. In clinical practice, the contextual structure of nodules and the accumulated experience of radiologists are the two core elements related to the accuracy of identification of benign and malignant nodules. Contextual information provides comprehensive information about nodules such as location, shape, and peripheral vessels, and experienced radiologists can search for clues from previous cases as a reference to enrich the basis of decision-making. In this paper, we propose a radiologist-inspired method to simulate the diagnostic process of radiologists, which is composed of context parsing and prototype recalling modules. The context parsing module first segments the context structure of nodules and then aggregates contextual information for a more comprehensive understanding of the nodule. The prototype recalling module utilizes prototype-based learning to condense previously learned cases as prototypes for comparative analysis, which is updated online in a momentum way during training. Building on the two modules, our method leverages both the intrinsic characteristics of the nodules and the external knowledge accumulated from other nodules to achieve a sound diagnosis. To meet the needs of both low-dose and noncontrast screening, we collect a large-scale dataset of 12,852 and 4,029 nodules from low-dose and noncontrast CTs respectively, each with pathology- or follow-up-confirmed labels. Experiments on several datasets demonstrate that our method achieves advanced screening performance on both low-dose and noncontrast scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge