Sinuo Wang
O-Researcher: An Open Ended Deep Research Model via Multi-Agent Distillation and Agentic RL
Jan 07, 2026Abstract:The performance gap between closed-source and open-source large language models (LLMs) is largely attributed to disparities in access to high-quality training data. To bridge this gap, we introduce a novel framework for the automated synthesis of sophisticated, research-grade instructional data. Our approach centers on a multi-agent workflow where collaborative AI agents simulate complex tool-integrated reasoning to generate diverse and high-fidelity data end-to-end. Leveraging this synthesized data, we develop a two-stage training strategy that integrates supervised fine-tuning with a novel reinforcement learning method, designed to maximize model alignment and capability. Extensive experiments demonstrate that our framework empowers open-source models across multiple scales, enabling them to achieve new state-of-the-art performance on the major deep research benchmark. This work provides a scalable and effective pathway for advancing open-source LLMs without relying on proprietary data or models.
Large-scale and Fine-grained Vision-language Pre-training for Enhanced CT Image Understanding
Jan 24, 2025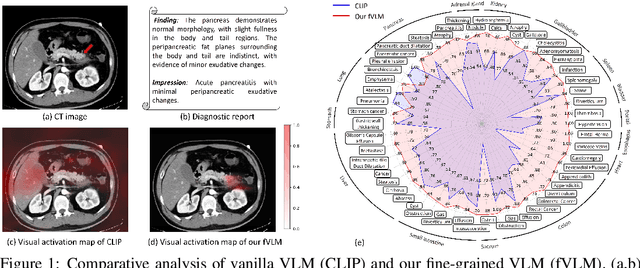
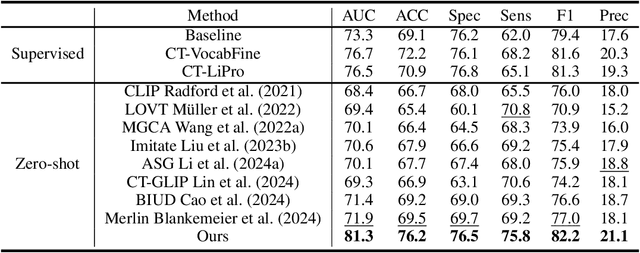
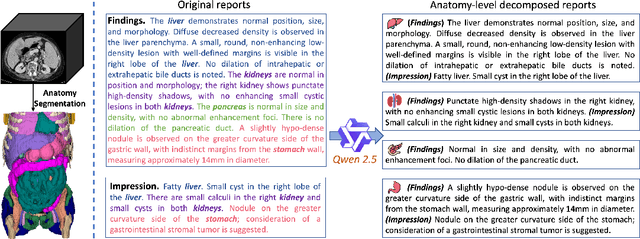
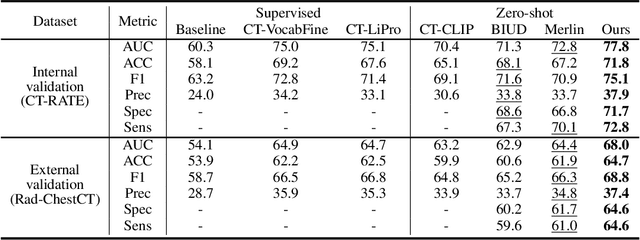
Abstract:Artificial intelligence (AI) shows great potential in assisting radiologists to improve the efficiency and accuracy of medical image interpretation and diagnosis. However, a versatile AI model requires large-scale data and comprehensive annotations, which are often impractical in medical settings. Recent studies leverage radiology reports as a naturally high-quality supervision for medical images, using contrastive language-image pre-training (CLIP) to develop language-informed models for radiological image interpretation. Nonetheless, these approaches typically contrast entire images with reports, neglecting the local associations between imaging regions and report sentences, which may undermine model performance and interoperability. In this paper, we propose a fine-grained vision-language model (fVLM) for anatomy-level CT image interpretation. Specifically, we explicitly match anatomical regions of CT images with corresponding descriptions in radiology reports and perform contrastive pre-training for each anatomy individually. Fine-grained alignment, however, faces considerable false-negative challenges, mainly from the abundance of anatomy-level healthy samples and similarly diseased abnormalities. To tackle this issue, we propose identifying false negatives of both normal and abnormal samples and calibrating contrastive learning from patient-level to disease-aware pairing. We curated the largest CT dataset to date, comprising imaging and report data from 69,086 patients, and conducted a comprehensive evaluation of 54 major and important disease diagnosis tasks across 15 main anatomies. Experimental results demonstrate the substantial potential of fVLM in versatile medical image interpretation. In the zero-shot classification task, we achieved an average AUC of 81.3% on 54 diagnosis tasks, surpassing CLIP and supervised methods by 12.9% and 8.0%, respectively.
A Survey of Medical Vision-and-Language Applications and Their Techniques
Nov 19, 2024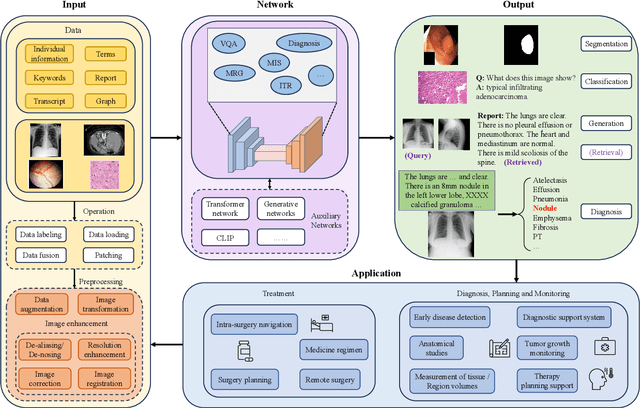
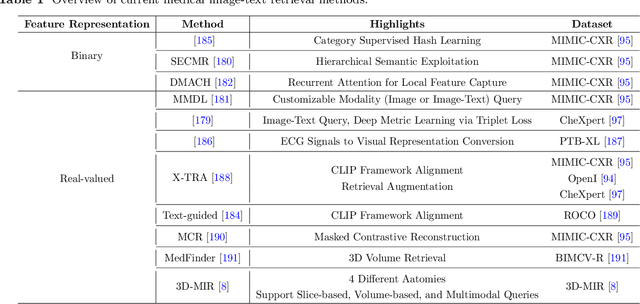
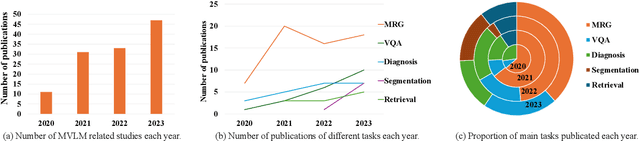
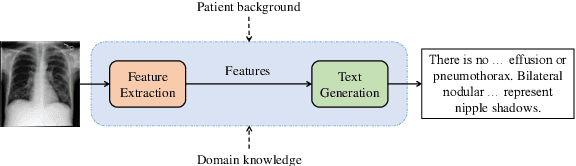
Abstract:Medical vision-and-language models (MVLMs) have attracted substantial interest due to their capability to offer a natural language interface for interpreting complex medical data. Their applications are versatile and have the potential to improve diagnostic accuracy and decision-making for individual patients while also contributing to enhanced public health monitoring, disease surveillance, and policy-making through more efficient analysis of large data sets. MVLMS integrate natural language processing with medical images to enable a more comprehensive and contextual understanding of medical images alongside their corresponding textual information. Unlike general vision-and-language models trained on diverse, non-specialized datasets, MVLMs are purpose-built for the medical domain, automatically extracting and interpreting critical information from medical images and textual reports to support clinical decision-making. Popular clinical applications of MVLMs include automated medical report generation, medical visual question answering, medical multimodal segmentation, diagnosis and prognosis and medical image-text retrieval. Here, we provide a comprehensive overview of MVLMs and the various medical tasks to which they have been applied. We conduct a detailed analysis of various vision-and-language model architectures, focusing on their distinct strategies for cross-modal integration/exploitation of medical visual and textual features. We also examine the datasets used for these tasks and compare the performance of different models based on standardized evaluation metrics. Furthermore, we highlight potential challenges and summarize future research trends and directions. The full collection of papers and codes is available at: https://github.com/YtongXie/Medical-Vision-and-Language-Tasks-and-Methodologies-A-Survey.
PairAug: What Can Augmented Image-Text Pairs Do for Radiology?
Apr 07, 2024



Abstract:Current vision-language pre-training (VLP) methodologies predominantly depend on paired image-text datasets, a resource that is challenging to acquire in radiology due to privacy considerations and labelling complexities. Data augmentation provides a practical solution to overcome the issue of data scarcity, however, most augmentation methods exhibit a limited focus, prioritising either image or text augmentation exclusively. Acknowledging this limitation, our objective is to devise a framework capable of concurrently augmenting medical image and text data. We design a Pairwise Augmentation (PairAug) approach that contains an Inter-patient Augmentation (InterAug) branch and an Intra-patient Augmentation (IntraAug) branch. Specifically, the InterAug branch of our approach generates radiology images using synthesised yet plausible reports derived from a Large Language Model (LLM). The generated pairs can be considered a collection of new patient cases since they are artificially created and may not exist in the original dataset. In contrast, the IntraAug branch uses newly generated reports to manipulate images. This process allows us to create new paired data for each individual with diverse medical conditions. Our extensive experiments on various downstream tasks covering medical image classification zero-shot and fine-tuning analysis demonstrate that our PairAug, concurrently expanding both image and text data, substantially outperforms image-/text-only expansion baselines and advanced medical VLP baselines. Our code is released at \url{https://github.com/YtongXie/PairAug}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge