Zhibin Liao
Seeing the Trees for the Forest: Rethinking Weakly-Supervised Medical Visual Grounding
May 21, 2025Abstract:Visual grounding (VG) is the capability to identify the specific regions in an image associated with a particular text description. In medical imaging, VG enhances interpretability by highlighting relevant pathological features corresponding to textual descriptions, improving model transparency and trustworthiness for wider adoption of deep learning models in clinical practice. Current models struggle to associate textual descriptions with disease regions due to inefficient attention mechanisms and a lack of fine-grained token representations. In this paper, we empirically demonstrate two key observations. First, current VLMs assign high norms to background tokens, diverting the model's attention from regions of disease. Second, the global tokens used for cross-modal learning are not representative of local disease tokens. This hampers identifying correlations between the text and disease tokens. To address this, we introduce simple, yet effective Disease-Aware Prompting (DAP) process, which uses the explainability map of a VLM to identify the appropriate image features. This simple strategy amplifies disease-relevant regions while suppressing background interference. Without any additional pixel-level annotations, DAP improves visual grounding accuracy by 20.74% compared to state-of-the-art methods across three major chest X-ray datasets.
Localizing Before Answering: A Hallucination Evaluation Benchmark for Grounded Medical Multimodal LLMs
May 05, 2025Abstract:Medical Large Multi-modal Models (LMMs) have demonstrated remarkable capabilities in medical data interpretation. However, these models frequently generate hallucinations contradicting source evidence, particularly due to inadequate localization reasoning. This work reveals a critical limitation in current medical LMMs: instead of analyzing relevant pathological regions, they often rely on linguistic patterns or attend to irrelevant image areas when responding to disease-related queries. To address this, we introduce HEAL-MedVQA (Hallucination Evaluation via Localization MedVQA), a comprehensive benchmark designed to evaluate LMMs' localization abilities and hallucination robustness. HEAL-MedVQA features (i) two innovative evaluation protocols to assess visual and textual shortcut learning, and (ii) a dataset of 67K VQA pairs, with doctor-annotated anatomical segmentation masks for pathological regions. To improve visual reasoning, we propose the Localize-before-Answer (LobA) framework, which trains LMMs to localize target regions of interest and self-prompt to emphasize segmented pathological areas, generating grounded and reliable answers. Experimental results demonstrate that our approach significantly outperforms state-of-the-art biomedical LMMs on the challenging HEAL-MedVQA benchmark, advancing robustness in medical VQA.
Interactive Medical Image Analysis with Concept-based Similarity Reasoning
Mar 11, 2025Abstract:The ability to interpret and intervene model decisions is important for the adoption of computer-aided diagnosis methods in clinical workflows. Recent concept-based methods link the model predictions with interpretable concepts and modify their activation scores to interact with the model. However, these concepts are at the image level, which hinders the model from pinpointing the exact patches the concepts are activated. Alternatively, prototype-based methods learn representations from training image patches and compare these with test image patches, using the similarity scores for final class prediction. However, interpreting the underlying concepts of these patches can be challenging and often necessitates post-hoc guesswork. To address this issue, this paper introduces the novel Concept-based Similarity Reasoning network (CSR), which offers (i) patch-level prototype with intrinsic concept interpretation, and (ii) spatial interactivity. First, the proposed CSR provides localized explanation by grounding prototypes of each concept on image regions. Second, our model introduces novel spatial-level interaction, allowing doctors to engage directly with specific image areas, making it an intuitive and transparent tool for medical imaging. CSR improves upon prior state-of-the-art interpretable methods by up to 4.5\% across three biomedical datasets. Our code is released at https://github.com/tadeephuy/InteractCSR.
* Accepted CVPR2025
ProjectedEx: Enhancing Generation in Explainable AI for Prostate Cancer
Jan 02, 2025



Abstract:Prostate cancer, a growing global health concern, necessitates precise diagnostic tools, with Magnetic Resonance Imaging (MRI) offering high-resolution soft tissue imaging that significantly enhances diagnostic accuracy. Recent advancements in explainable AI and representation learning have significantly improved prostate cancer diagnosis by enabling automated and precise lesion classification. However, existing explainable AI methods, particularly those based on frameworks like generative adversarial networks (GANs), are predominantly developed for natural image generation, and their application to medical imaging often leads to suboptimal performance due to the unique characteristics and complexity of medical image. To address these challenges, our paper introduces three key contributions. First, we propose ProjectedEx, a generative framework that provides interpretable, multi-attribute explanations, effectively linking medical image features to classifier decisions. Second, we enhance the encoder module by incorporating feature pyramids, which enables multiscale feedback to refine the latent space and improves the quality of generated explanations. Additionally, we conduct comprehensive experiments on both the generator and classifier, demonstrating the clinical relevance and effectiveness of ProjectedEx in enhancing interpretability and supporting the adoption of AI in medical settings. Code will be released at https://github.com/Richardqiyi/ProjectedEx
A Survey of Medical Vision-and-Language Applications and Their Techniques
Nov 19, 2024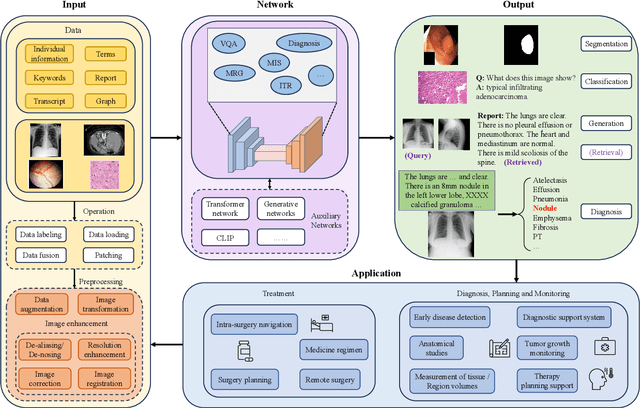
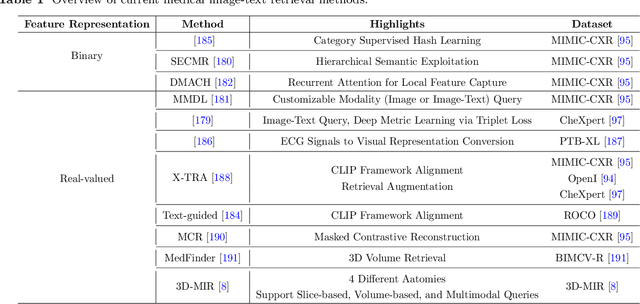
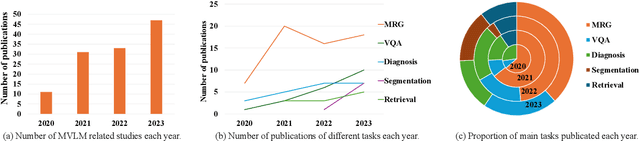
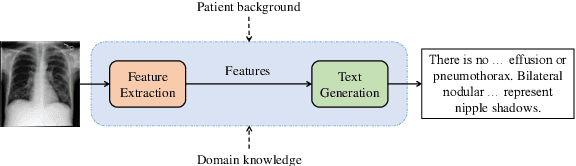
Abstract:Medical vision-and-language models (MVLMs) have attracted substantial interest due to their capability to offer a natural language interface for interpreting complex medical data. Their applications are versatile and have the potential to improve diagnostic accuracy and decision-making for individual patients while also contributing to enhanced public health monitoring, disease surveillance, and policy-making through more efficient analysis of large data sets. MVLMS integrate natural language processing with medical images to enable a more comprehensive and contextual understanding of medical images alongside their corresponding textual information. Unlike general vision-and-language models trained on diverse, non-specialized datasets, MVLMs are purpose-built for the medical domain, automatically extracting and interpreting critical information from medical images and textual reports to support clinical decision-making. Popular clinical applications of MVLMs include automated medical report generation, medical visual question answering, medical multimodal segmentation, diagnosis and prognosis and medical image-text retrieval. Here, we provide a comprehensive overview of MVLMs and the various medical tasks to which they have been applied. We conduct a detailed analysis of various vision-and-language model architectures, focusing on their distinct strategies for cross-modal integration/exploitation of medical visual and textual features. We also examine the datasets used for these tasks and compare the performance of different models based on standardized evaluation metrics. Furthermore, we highlight potential challenges and summarize future research trends and directions. The full collection of papers and codes is available at: https://github.com/YtongXie/Medical-Vision-and-Language-Tasks-and-Methodologies-A-Survey.
MSDet: Receptive Field Enhanced Multiscale Detection for Tiny Pulmonary Nodule
Sep 21, 2024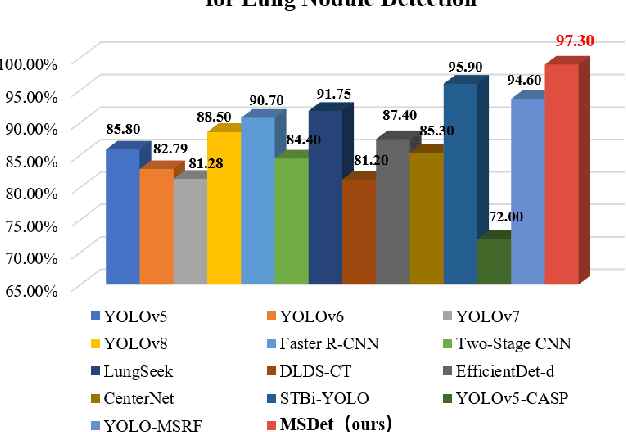
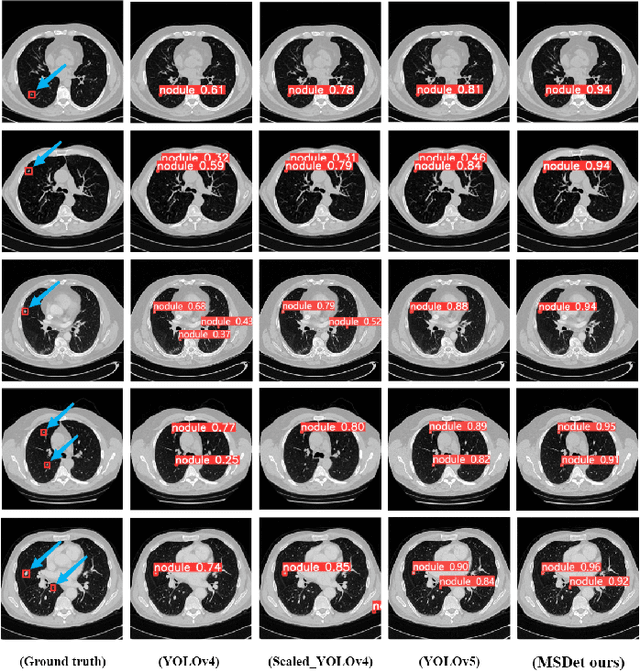
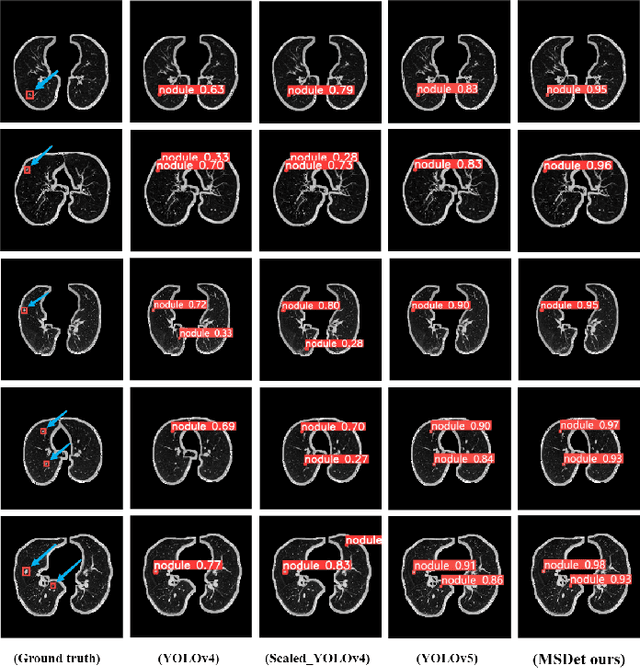
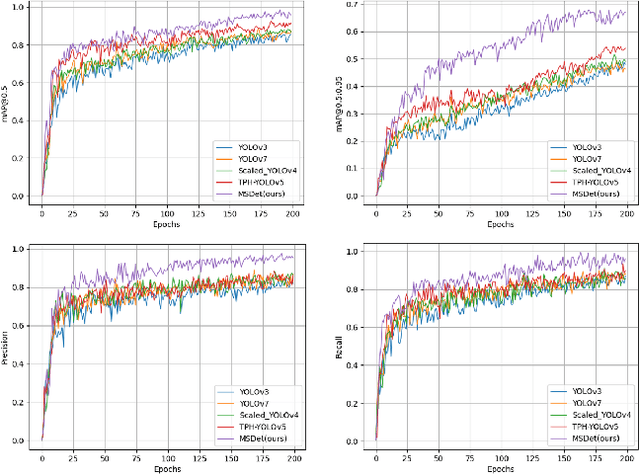
Abstract:Pulmonary nodules are critical indicators for the early diagnosis of lung cancer, making their detection essential for timely treatment. However, traditional CT imaging methods suffered from cumbersome procedures, low detection rates, and poor localization accuracy. The subtle differences between pulmonary nodules and surrounding tissues in complex lung CT images, combined with repeated downsampling in feature extraction networks, often lead to missed or false detections of small nodules. Existing methods such as FPN, with its fixed feature fusion and limited receptive field, struggle to effectively overcome these issues. To address these challenges, our paper proposed three key contributions: Firstly, we proposed MSDet, a multiscale attention and receptive field network for detecting tiny pulmonary nodules. Secondly, we proposed the extended receptive domain (ERD) strategy to capture richer contextual information and reduce false positives caused by nodule occlusion. We also proposed the position channel attention mechanism (PCAM) to optimize feature learning and reduce multiscale detection errors, and designed the tiny object detection block (TODB) to enhance the detection of tiny nodules. Lastly, we conducted thorough experiments on the public LUNA16 dataset, achieving state-of-the-art performance, with an mAP improvement of 8.8% over the previous state-of-the-art method YOLOv8. These advancements significantly boosted detection accuracy and reliability, providing a more effective solution for early lung cancer diagnosis. The code will be available at https://github.com/CaiGuoHui123/MSDet
AdaCBM: An Adaptive Concept Bottleneck Model for Explainable and Accurate Diagnosis
Aug 04, 2024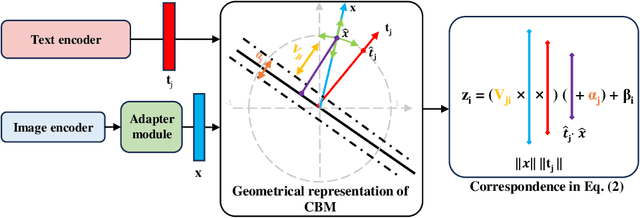
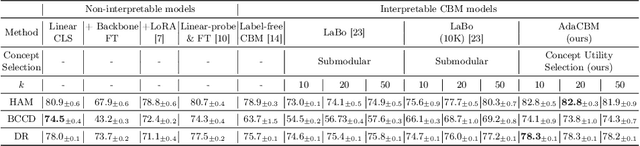
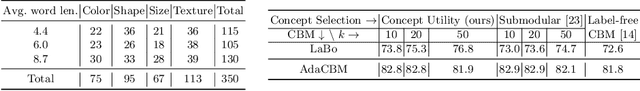
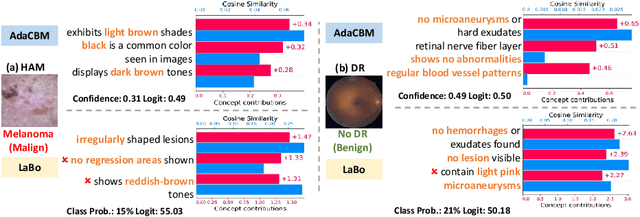
Abstract:The integration of vision-language models such as CLIP and Concept Bottleneck Models (CBMs) offers a promising approach to explaining deep neural network (DNN) decisions using concepts understandable by humans, addressing the black-box concern of DNNs. While CLIP provides both explainability and zero-shot classification capability, its pre-training on generic image and text data may limit its classification accuracy and applicability to medical image diagnostic tasks, creating a transfer learning problem. To maintain explainability and address transfer learning needs, CBM methods commonly design post-processing modules after the bottleneck module. However, this way has been ineffective. This paper takes an unconventional approach by re-examining the CBM framework through the lens of its geometrical representation as a simple linear classification system. The analysis uncovers that post-CBM fine-tuning modules merely rescale and shift the classification outcome of the system, failing to fully leverage the system's learning potential. We introduce an adaptive module strategically positioned between CLIP and CBM to bridge the gap between source and downstream domains. This simple yet effective approach enhances classification performance while preserving the explainability afforded by the framework. Our work offers a comprehensive solution that encompasses the entire process, from concept discovery to model training, providing a holistic recipe for leveraging the strengths of GPT, CLIP, and CBM.
Structural Attention: Rethinking Transformer for Unpaired Medical Image Synthesis
Jun 27, 2024



Abstract:Unpaired medical image synthesis aims to provide complementary information for an accurate clinical diagnostics, and address challenges in obtaining aligned multi-modal medical scans. Transformer-based models excel in imaging translation tasks thanks to their ability to capture long-range dependencies. Although effective in supervised training settings, their performance falters in unpaired image synthesis, particularly in synthesizing structural details. This paper empirically demonstrates that, lacking strong inductive biases, Transformer can converge to non-optimal solutions in the absence of paired data. To address this, we introduce UNet Structured Transformer (UNest), a novel architecture incorporating structural inductive biases for unpaired medical image synthesis. We leverage the foundational Segment-Anything Model to precisely extract the foreground structure and perform structural attention within the main anatomy. This guides the model to learn key anatomical regions, thus improving structural synthesis under the lack of supervision in unpaired training. Evaluated on two public datasets, spanning three modalities, i.e., MR, CT, and PET, UNest improves recent methods by up to 19.30% across six medical image synthesis tasks. Our code is released at https://github.com/HieuPhan33/MICCAI2024-UNest.
JointViT: Modeling Oxygen Saturation Levels with Joint Supervision on Long-Tailed OCTA
Apr 18, 2024Abstract:The oxygen saturation level in the blood (SaO2) is crucial for health, particularly in relation to sleep-related breathing disorders. However, continuous monitoring of SaO2 is time-consuming and highly variable depending on patients' conditions. Recently, optical coherence tomography angiography (OCTA) has shown promising development in rapidly and effectively screening eye-related lesions, offering the potential for diagnosing sleep-related disorders. To bridge this gap, our paper presents three key contributions. Firstly, we propose JointViT, a novel model based on the Vision Transformer architecture, incorporating a joint loss function for supervision. Secondly, we introduce a balancing augmentation technique during data preprocessing to improve the model's performance, particularly on the long-tail distribution within the OCTA dataset. Lastly, through comprehensive experiments on the OCTA dataset, our proposed method significantly outperforms other state-of-the-art methods, achieving improvements of up to 12.28% in overall accuracy. This advancement lays the groundwork for the future utilization of OCTA in diagnosing sleep-related disorders. See project website https://steve-zeyu-zhang.github.io/JointViT
CAPE: CAM as a Probabilistic Ensemble for Enhanced DNN Interpretation
Apr 04, 2024Abstract:Deep Neural Networks (DNNs) are widely used for visual classification tasks, but their complex computation process and black-box nature hinder decision transparency and interpretability. Class activation maps (CAMs) and recent variants provide ways to visually explain the DNN decision-making process by displaying 'attention' heatmaps of the DNNs. Nevertheless, the CAM explanation only offers relative attention information, that is, on an attention heatmap, we can interpret which image region is more or less important than the others. However, these regions cannot be meaningfully compared across classes, and the contribution of each region to the model's class prediction is not revealed. To address these challenges that ultimately lead to better DNN Interpretation, in this paper, we propose CAPE, a novel reformulation of CAM that provides a unified and probabilistically meaningful assessment of the contributions of image regions. We quantitatively and qualitatively compare CAPE with state-of-the-art CAM methods on CUB and ImageNet benchmark datasets to demonstrate enhanced interpretability. We also test on a cytology imaging dataset depicting a challenging Chronic Myelomonocytic Leukemia (CMML) diagnosis problem. Code is available at: https://github.com/AIML-MED/CAPE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge