Johan W. Verjans
Seeing the Trees for the Forest: Rethinking Weakly-Supervised Medical Visual Grounding
May 21, 2025Abstract:Visual grounding (VG) is the capability to identify the specific regions in an image associated with a particular text description. In medical imaging, VG enhances interpretability by highlighting relevant pathological features corresponding to textual descriptions, improving model transparency and trustworthiness for wider adoption of deep learning models in clinical practice. Current models struggle to associate textual descriptions with disease regions due to inefficient attention mechanisms and a lack of fine-grained token representations. In this paper, we empirically demonstrate two key observations. First, current VLMs assign high norms to background tokens, diverting the model's attention from regions of disease. Second, the global tokens used for cross-modal learning are not representative of local disease tokens. This hampers identifying correlations between the text and disease tokens. To address this, we introduce simple, yet effective Disease-Aware Prompting (DAP) process, which uses the explainability map of a VLM to identify the appropriate image features. This simple strategy amplifies disease-relevant regions while suppressing background interference. Without any additional pixel-level annotations, DAP improves visual grounding accuracy by 20.74% compared to state-of-the-art methods across three major chest X-ray datasets.
Interactive Medical Image Analysis with Concept-based Similarity Reasoning
Mar 11, 2025Abstract:The ability to interpret and intervene model decisions is important for the adoption of computer-aided diagnosis methods in clinical workflows. Recent concept-based methods link the model predictions with interpretable concepts and modify their activation scores to interact with the model. However, these concepts are at the image level, which hinders the model from pinpointing the exact patches the concepts are activated. Alternatively, prototype-based methods learn representations from training image patches and compare these with test image patches, using the similarity scores for final class prediction. However, interpreting the underlying concepts of these patches can be challenging and often necessitates post-hoc guesswork. To address this issue, this paper introduces the novel Concept-based Similarity Reasoning network (CSR), which offers (i) patch-level prototype with intrinsic concept interpretation, and (ii) spatial interactivity. First, the proposed CSR provides localized explanation by grounding prototypes of each concept on image regions. Second, our model introduces novel spatial-level interaction, allowing doctors to engage directly with specific image areas, making it an intuitive and transparent tool for medical imaging. CSR improves upon prior state-of-the-art interpretable methods by up to 4.5\% across three biomedical datasets. Our code is released at https://github.com/tadeephuy/InteractCSR.
* Accepted CVPR2025
AdaCBM: An Adaptive Concept Bottleneck Model for Explainable and Accurate Diagnosis
Aug 04, 2024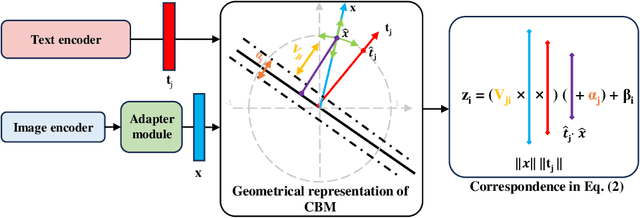
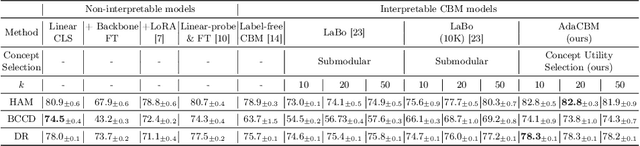
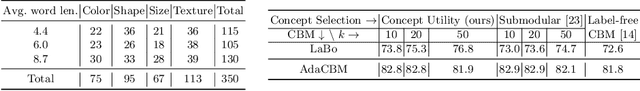
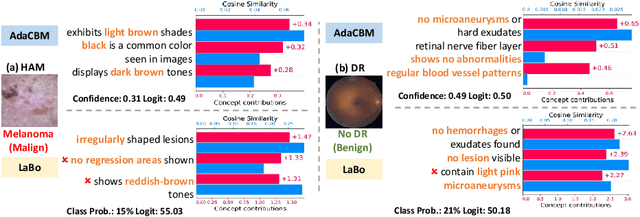
Abstract:The integration of vision-language models such as CLIP and Concept Bottleneck Models (CBMs) offers a promising approach to explaining deep neural network (DNN) decisions using concepts understandable by humans, addressing the black-box concern of DNNs. While CLIP provides both explainability and zero-shot classification capability, its pre-training on generic image and text data may limit its classification accuracy and applicability to medical image diagnostic tasks, creating a transfer learning problem. To maintain explainability and address transfer learning needs, CBM methods commonly design post-processing modules after the bottleneck module. However, this way has been ineffective. This paper takes an unconventional approach by re-examining the CBM framework through the lens of its geometrical representation as a simple linear classification system. The analysis uncovers that post-CBM fine-tuning modules merely rescale and shift the classification outcome of the system, failing to fully leverage the system's learning potential. We introduce an adaptive module strategically positioned between CLIP and CBM to bridge the gap between source and downstream domains. This simple yet effective approach enhances classification performance while preserving the explainability afforded by the framework. Our work offers a comprehensive solution that encompasses the entire process, from concept discovery to model training, providing a holistic recipe for leveraging the strengths of GPT, CLIP, and CBM.
Structural Attention: Rethinking Transformer for Unpaired Medical Image Synthesis
Jun 27, 2024



Abstract:Unpaired medical image synthesis aims to provide complementary information for an accurate clinical diagnostics, and address challenges in obtaining aligned multi-modal medical scans. Transformer-based models excel in imaging translation tasks thanks to their ability to capture long-range dependencies. Although effective in supervised training settings, their performance falters in unpaired image synthesis, particularly in synthesizing structural details. This paper empirically demonstrates that, lacking strong inductive biases, Transformer can converge to non-optimal solutions in the absence of paired data. To address this, we introduce UNet Structured Transformer (UNest), a novel architecture incorporating structural inductive biases for unpaired medical image synthesis. We leverage the foundational Segment-Anything Model to precisely extract the foreground structure and perform structural attention within the main anatomy. This guides the model to learn key anatomical regions, thus improving structural synthesis under the lack of supervision in unpaired training. Evaluated on two public datasets, spanning three modalities, i.e., MR, CT, and PET, UNest improves recent methods by up to 19.30% across six medical image synthesis tasks. Our code is released at https://github.com/HieuPhan33/MICCAI2024-UNest.
CAPE: CAM as a Probabilistic Ensemble for Enhanced DNN Interpretation
Apr 04, 2024Abstract:Deep Neural Networks (DNNs) are widely used for visual classification tasks, but their complex computation process and black-box nature hinder decision transparency and interpretability. Class activation maps (CAMs) and recent variants provide ways to visually explain the DNN decision-making process by displaying 'attention' heatmaps of the DNNs. Nevertheless, the CAM explanation only offers relative attention information, that is, on an attention heatmap, we can interpret which image region is more or less important than the others. However, these regions cannot be meaningfully compared across classes, and the contribution of each region to the model's class prediction is not revealed. To address these challenges that ultimately lead to better DNN Interpretation, in this paper, we propose CAPE, a novel reformulation of CAM that provides a unified and probabilistically meaningful assessment of the contributions of image regions. We quantitatively and qualitatively compare CAPE with state-of-the-art CAM methods on CUB and ImageNet benchmark datasets to demonstrate enhanced interpretability. We also test on a cytology imaging dataset depicting a challenging Chronic Myelomonocytic Leukemia (CMML) diagnosis problem. Code is available at: https://github.com/AIML-MED/CAPE.
Decomposing Disease Descriptions for Enhanced Pathology Detection: A Multi-Aspect Vision-Language Matching Framework
Mar 12, 2024



Abstract:Medical vision language pre-training (VLP) has emerged as a frontier of research, enabling zero-shot pathological recognition by comparing the query image with the textual descriptions for each disease. Due to the complex semantics of biomedical texts, current methods struggle to align medical images with key pathological findings in unstructured reports. This leads to the misalignment with the target disease's textual representation. In this paper, we introduce a novel VLP framework designed to dissect disease descriptions into their fundamental aspects, leveraging prior knowledge about the visual manifestations of pathologies. This is achieved by consulting a large language model and medical experts. Integrating a Transformer module, our approach aligns an input image with the diverse elements of a disease, generating aspect-centric image representations. By consolidating the matches from each aspect, we improve the compatibility between an image and its associated disease. Additionally, capitalizing on the aspect-oriented representations, we present a dual-head Transformer tailored to process known and unknown diseases, optimizing the comprehensive detection efficacy. Conducting experiments on seven downstream datasets, ours outperforms recent methods by up to 8.07% and 11.23% in AUC scores for seen and novel categories, respectively. Our code is released at \href{https://github.com/HieuPhan33/MAVL}{https://github.com/HieuPhan33/MAVL}.
Structure-Preserving Synthesis: MaskGAN for Unpaired MR-CT Translation
Aug 01, 2023



Abstract:Medical image synthesis is a challenging task due to the scarcity of paired data. Several methods have applied CycleGAN to leverage unpaired data, but they often generate inaccurate mappings that shift the anatomy. This problem is further exacerbated when the images from the source and target modalities are heavily misaligned. Recently, current methods have aimed to address this issue by incorporating a supplementary segmentation network. Unfortunately, this strategy requires costly and time-consuming pixel-level annotations. To overcome this problem, this paper proposes MaskGAN, a novel and cost-effective framework that enforces structural consistency by utilizing automatically extracted coarse masks. Our approach employs a mask generator to outline anatomical structures and a content generator to synthesize CT contents that align with these structures. Extensive experiments demonstrate that MaskGAN outperforms state-of-the-art synthesis methods on a challenging pediatric dataset, where MR and CT scans are heavily misaligned due to rapid growth in children. Specifically, MaskGAN excels in preserving anatomical structures without the need for expert annotations. The code for this paper can be found at https://github.com/HieuPhan33/MaskGAN.
* Accepted to MICCAI 2023
CNN Attention Guidance for Improved Orthopedics Radiographic Fracture Classification
Mar 21, 2022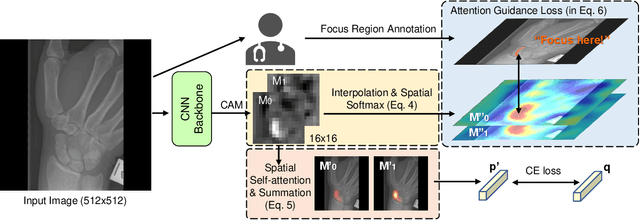
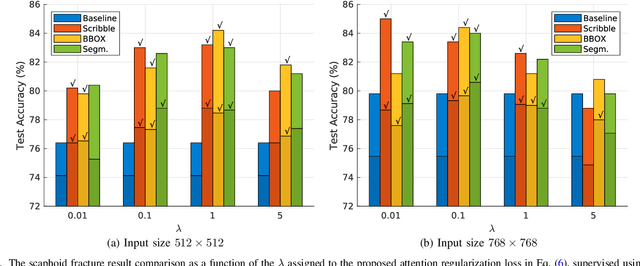
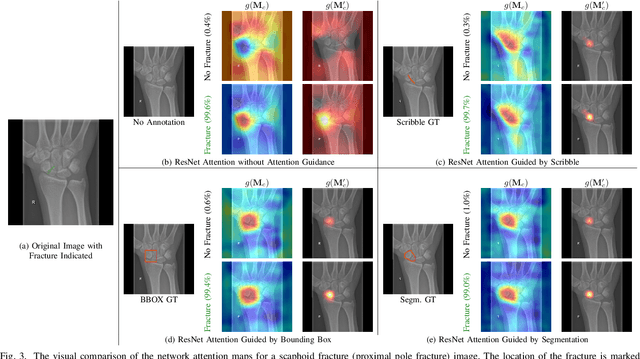
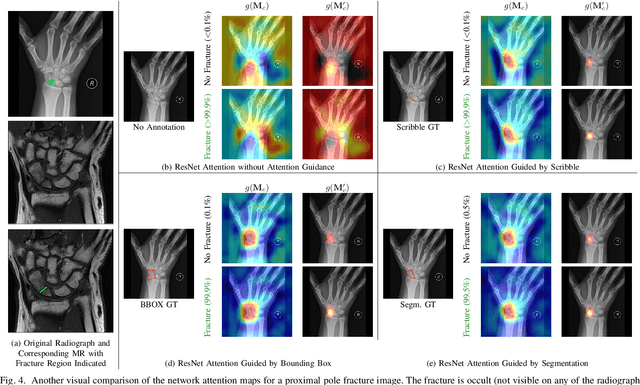
Abstract:Convolutional neural networks (CNNs) have gained significant popularity in orthopedic imaging in recent years due to their ability to solve fracture classification problems. A common criticism of CNNs is their opaque learning and reasoning process, making it difficult to trust machine diagnosis and the subsequent adoption of such algorithms in clinical setting. This is especially true when the CNN is trained with limited amount of medical data, which is a common issue as curating sufficiently large amount of annotated medical imaging data is a long and costly process. While interest has been devoted to explaining CNN learnt knowledge by visualizing network attention, the utilization of the visualized attention to improve network learning has been rarely investigated. This paper explores the effectiveness of regularizing CNN network with human-provided attention guidance on where in the image the network should look for answering clues. On two orthopedics radiographic fracture classification datasets, through extensive experiments we demonstrate that explicit human-guided attention indeed can direct correct network attention and consequently significantly improve classification performance. The development code for the proposed attention guidance is publicly available on GitHub.
Multi-centred Strong Augmentation via Contrastive Learning for Unsupervised Lesion Detection and Segmentation
Sep 03, 2021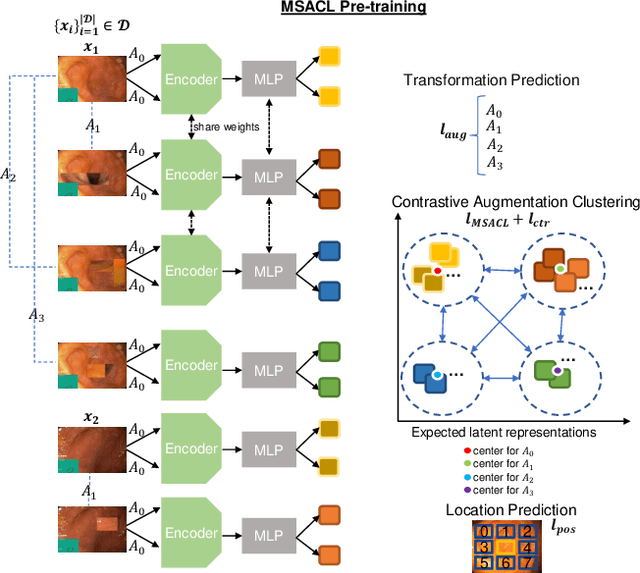
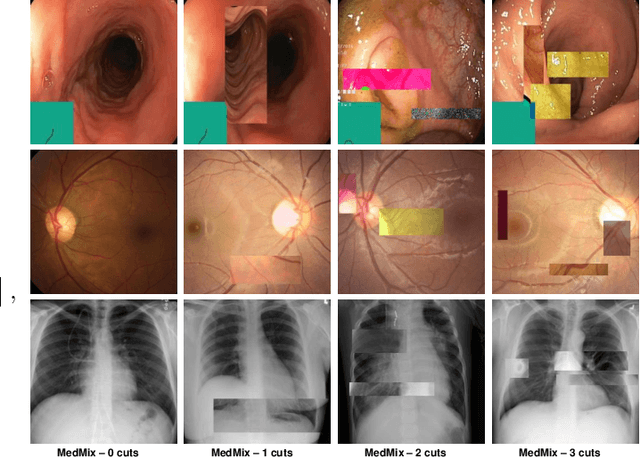
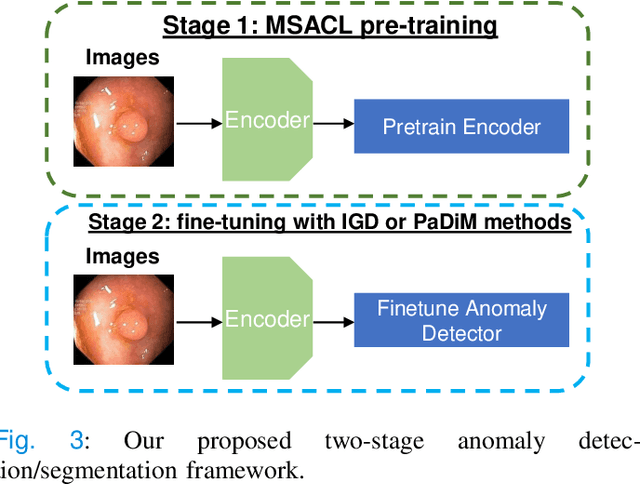
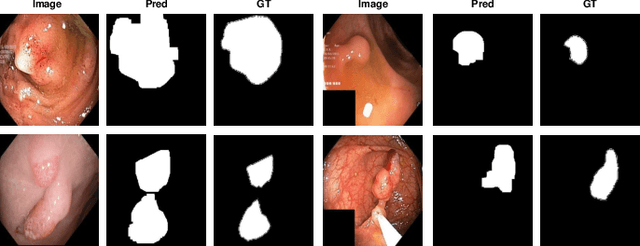
Abstract:The scarcity of high quality medical image annotations hinders the implementation of accurate clinical applications for detecting and segmenting abnormal lesions. To mitigate this issue, the scientific community is working on the development of unsupervised anomaly detection (UAD) systems that learn from a training set containing only normal (i.e., healthy) images, where abnormal samples (i.e., unhealthy) are detected and segmented based on how much they deviate from the learned distribution of normal samples. One significant challenge faced by UAD methods is how to learn effective low-dimensional image representations that are sensitive enough to detect and segment abnormal lesions of varying size, appearance and shape. To address this challenge, we propose a novel self-supervised UAD pre-training algorithm, named Multi-centred Strong Augmentation via Contrastive Learning (MSACL). MSACL learns representations by separating several types of strong and weak augmentations of normal image samples, where the weak augmentations represent normal images and strong augmentations denote synthetic abnormal images. To produce such strong augmentations, we introduce MedMix, a novel data augmentation strategy that creates new training images with realistic looking lesions (i.e., anomalies) in normal images. The pre-trained representations from MSACL are generic and can be used to improve the efficacy of different types of off-the-shelf state-of-the-art (SOTA) UAD models. Comprehensive experimental results show that the use of MSACL largely improves these SOTA UAD models on four medical imaging datasets from diverse organs, namely colonoscopy, fundus screening and covid-19 chest-ray datasets.
Constrained Contrastive Distribution Learning for Unsupervised Anomaly Detection and Localisation in Medical Images
Mar 05, 2021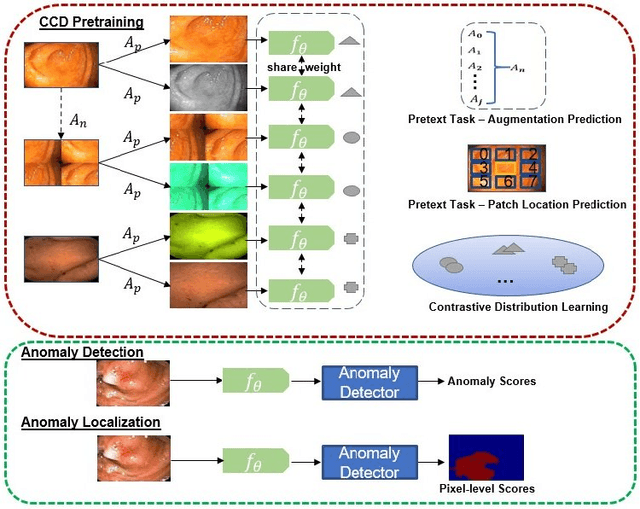
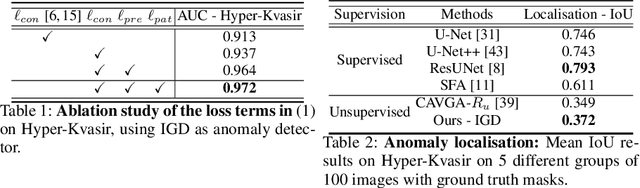
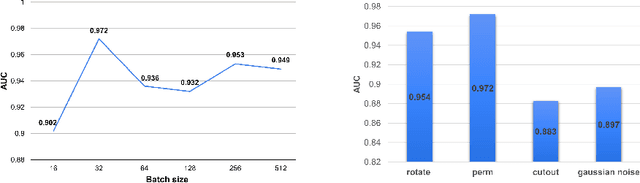
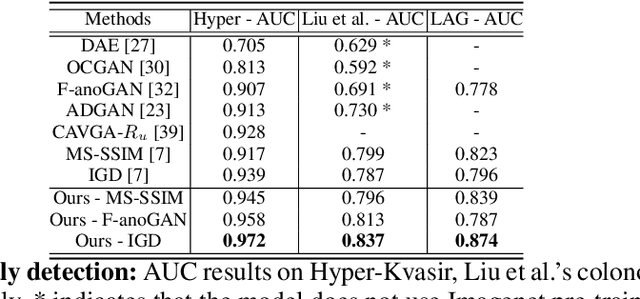
Abstract:Unsupervised anomaly detection (UAD) learns one-class classifiers exclusively with normal (i.e., healthy) images to detect any abnormal (i.e., unhealthy) samples that do not conform to the expected normal patterns. UAD has two main advantages over its fully supervised counterpart. Firstly, it is able to directly leverage large datasets available from health screening programs that contain mostly normal image samples, avoiding the costly manual labelling of abnormal samples and the subsequent issues involved in training with extremely class-imbalanced data. Further, UAD approaches can potentially detect and localise any type of lesions that deviate from the normal patterns. One significant challenge faced by UAD methods is how to learn effective low-dimensional image representations to detect and localise subtle abnormalities, generally consisting of small lesions. To address this challenge, we propose a novel self-supervised representation learning method, called Constrained Contrastive Distribution learning for anomaly detection (CCD), which learns fine-grained feature representations by simultaneously predicting the distribution of augmented data and image contexts using contrastive learning with pretext constraints. The learned representations can be leveraged to train more anomaly-sensitive detection models. Extensive experiment results show that our method outperforms current state-of-the-art UAD approaches on three different colonoscopy and fundus screening datasets. Our code is available at https://github.com/tianyu0207/CCD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge