Yong Xia
SegRap2025: A Benchmark of Gross Tumor Volume and Lymph Node Clinical Target Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Jan 28, 2026Abstract:Accurate delineation of Gross Tumor Volume (GTV), Lymph Node Clinical Target Volume (LN CTV), and Organ-at-Risk (OAR) from Computed Tomography (CT) scans is essential for precise radiotherapy planning in Nasopharyngeal Carcinoma (NPC). Building upon SegRap2023, which focused on OAR and GTV segmentation using single-center paired non-contrast CT (ncCT) and contrast-enhanced CT (ceCT) scans, the SegRap2025 challenge aims to enhance the generalizability and robustness of segmentation models across imaging centers and modalities. SegRap2025 comprises two tasks: Task01 addresses GTV segmentation using paired CT from the SegRap2023 dataset, with an additional external testing set to evaluate cross-center generalization, and Task02 focuses on LN CTV segmentation using multi-center training data and an unseen external testing set, where each case contains paired CT scans or a single modality, emphasizing both cross-center and cross-modality robustness. This paper presents the challenge setup and provides a comprehensive analysis of the solutions submitted by ten participating teams. For GTV segmentation task, the top-performing models achieved average Dice Similarity Coefficient (DSC) of 74.61% and 56.79% on the internal and external testing cohorts, respectively. For LN CTV segmentation task, the highest average DSC values reached 60.24%, 60.50%, and 57.23% on paired CT, ceCT-only, and ncCT-only subsets, respectively. SegRap2025 establishes a large-scale multi-center, multi-modality benchmark for evaluating the generalization and robustness in radiotherapy target segmentation, providing valuable insights toward clinically applicable automated radiotherapy planning systems. The benchmark is available at: https://hilab-git.github.io/SegRap2025_Challenge.
V-Loop: Visual Logical Loop Verification for Hallucination Detection in Medical Visual Question Answering
Jan 26, 2026Abstract:Multimodal Large Language Models (MLLMs) have shown remarkable capability in assisting disease diagnosis in medical visual question answering (VQA). However, their outputs remain vulnerable to hallucinations (i.e., responses that contradict visual facts), posing significant risks in high-stakes medical scenarios. Recent introspective detection methods, particularly uncertainty-based approaches, offer computational efficiency but are fundamentally indirect, as they estimate predictive uncertainty for an image-question pair rather than verifying the factual correctness of a specific answer. To address this limitation, we propose Visual Logical Loop Verification (V-Loop), a training-free and plug-and-play framework for hallucination detection in medical VQA. V-Loop introduces a bidirectional reasoning process that forms a visually grounded logical loop to verify factual correctness. Given an input, the MLLM produces an answer for the primary input pair. V-Loop extracts semantic units from the primary QA pair, generates a verification question by conditioning on the answer unit to re-query the question unit, and enforces visual attention consistency to ensure answering both primary question and verification question rely on the same image evidence. If the verification answer matches the expected semantic content, the logical loop closes, indicating factual grounding; otherwise, the primary answer is flagged as hallucinated. Extensive experiments on multiple medical VQA benchmarks and MLLMs show that V-Loop consistently outperforms existing introspective methods, remains highly efficient, and further boosts uncertainty-based approaches when used in combination.
PAINT: Pathology-Aware Integrated Next-Scale Transformation for Virtual Immunohistochemistry
Jan 22, 2026Abstract:Virtual immunohistochemistry (IHC) aims to computationally synthesize molecular staining patterns from routine Hematoxylin and Eosin (H\&E) images, offering a cost-effective and tissue-efficient alternative to traditional physical staining. However, this task is particularly challenging: H\&E morphology provides ambiguous cues about protein expression, and similar tissue structures may correspond to distinct molecular states. Most existing methods focus on direct appearance synthesis to implicitly achieve cross-modal generation, often resulting in semantic inconsistencies due to insufficient structural priors. In this paper, we propose Pathology-Aware Integrated Next-Scale Transformation (PAINT), a visual autoregressive framework that reformulates the synthesis process as a structure-first conditional generation task. Unlike direct image translation, PAINT enforces a causal order by resolving molecular details conditioned on a global structural layout. Central to this approach is the introduction of a Spatial Structural Start Map (3S-Map), which grounds the autoregressive initialization in observed morphology, ensuring deterministic, spatially aligned synthesis. Experiments on the IHC4BC and MIST datasets demonstrate that PAINT outperforms state-of-the-art methods in structural fidelity and clinical downstream tasks, validating the potential of structure-guided autoregressive modeling.
FedBiCross: A Bi-Level Optimization Framework to Tackle Non-IID Challenges in Data-Free One-Shot Federated Learning on Medical Data
Jan 05, 2026Abstract:Data-free knowledge distillation-based one-shot federated learning (OSFL) trains a model in a single communication round without sharing raw data, making OSFL attractive for privacy-sensitive medical applications. However, existing methods aggregate predictions from all clients to form a global teacher. Under non-IID data, conflicting predictions cancel out during averaging, yielding near-uniform soft labels that provide weak supervision for distillation. We propose FedBiCross, a personalized OSFL framework with three stages: (1) clustering clients by model output similarity to form coherent sub-ensembles, (2) bi-level cross-cluster optimization that learns adaptive weights to selectively leverage beneficial cross-cluster knowledge while suppressing negative transfer, and (3) personalized distillation for client-specific adaptation. Experiments on four medical image datasets demonstrate that FedBiCross consistently outperforms state-of-the-art baselines across different non-IID degrees.
ECG-aBcDe: Overcoming Model Dependence, Encoding ECG into a Universal Language for Any LLM
Sep 16, 2025Abstract:Large Language Models (LLMs) hold significant promise for electrocardiogram (ECG) analysis, yet challenges remain regarding transferability, time-scale information learning, and interpretability. Current methods suffer from model-specific ECG encoders, hindering transfer across LLMs. Furthermore, LLMs struggle to capture crucial time-scale information inherent in ECGs due to Transformer limitations. And their black-box nature limits clinical adoption. To address these limitations, we introduce ECG-aBcDe, a novel ECG encoding method that transforms ECG signals into a universal ECG language readily interpretable by any LLM. By constructing a hybrid dataset of ECG language and natural language, ECG-aBcDe enables direct fine-tuning of pre-trained LLMs without architectural modifications, achieving "construct once, use anywhere" capability. Moreover, the bidirectional convertibility between ECG and ECG language of ECG-aBcDe allows for extracting attention heatmaps from ECG signals, significantly enhancing interpretability. Finally, ECG-aBcDe explicitly represents time-scale information, mitigating Transformer limitations. This work presents a new paradigm for integrating ECG analysis with LLMs. Compared with existing methods, our method achieves competitive performance on ROUGE-L and METEOR. Notably, it delivers significant improvements in the BLEU-4, with improvements of 2.8 times and 3.9 times in in-dataset and cross-dataset evaluations, respectively, reaching scores of 42.58 and 30.76. These results provide strong evidence for the feasibility of the new paradigm.
Unified Start, Personalized End: Progressive Pruning for Efficient 3D Medical Image Segmentation
Sep 11, 2025Abstract:3D medical image segmentation often faces heavy resource and time consumption, limiting its scalability and rapid deployment in clinical environments. Existing efficient segmentation models are typically static and manually designed prior to training, which restricts their adaptability across diverse tasks and makes it difficult to balance performance with resource efficiency. In this paper, we propose PSP-Seg, a progressive pruning framework that enables dynamic and efficient 3D segmentation. PSP-Seg begins with a redundant model and iteratively prunes redundant modules through a combination of block-wise pruning and a functional decoupling loss. We evaluate PSP-Seg on five public datasets, benchmarking it against seven state-of-the-art models and six efficient segmentation models. Results demonstrate that the lightweight variant, PSP-Seg-S, achieves performance on par with nnU-Net while reducing GPU memory usage by 42-45%, training time by 29-48%, and parameter number by 83-87% across all datasets. These findings underscore PSP-Seg's potential as a cost-effective yet high-performing alternative for widespread clinical application.
Toward Robust Medical Fairness: Debiased Dual-Modal Alignment via Text-Guided Attribute-Disentangled Prompt Learning for Vision-Language Models
Aug 26, 2025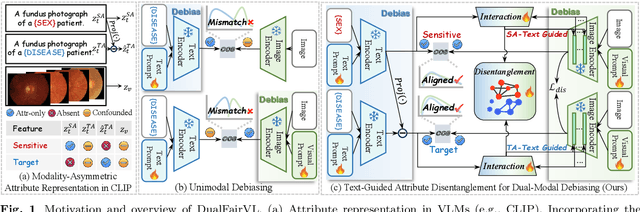
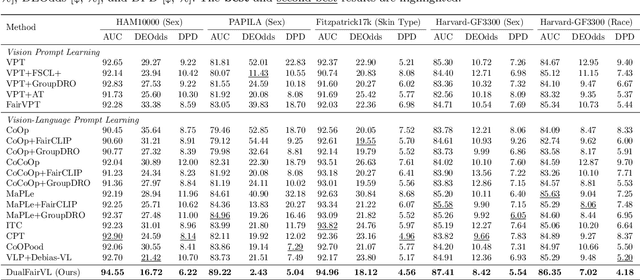
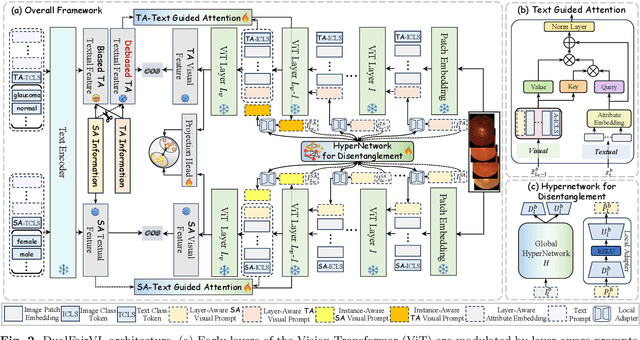
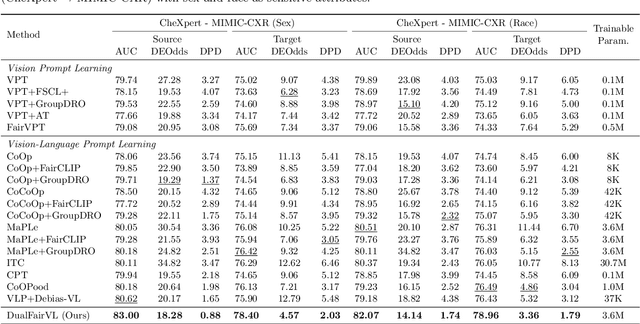
Abstract:Ensuring fairness across demographic groups in medical diagnosis is essential for equitable healthcare, particularly under distribution shifts caused by variations in imaging equipment and clinical practice. Vision-language models (VLMs) exhibit strong generalization, and text prompts encode identity attributes, enabling explicit identification and removal of sensitive directions. However, existing debiasing approaches typically address vision and text modalities independently, leaving residual cross-modal misalignment and fairness gaps. To address this challenge, we propose DualFairVL, a multimodal prompt-learning framework that jointly debiases and aligns cross-modal representations. DualFairVL employs a parallel dual-branch architecture that separates sensitive and target attributes, enabling disentangled yet aligned representations across modalities. Approximately orthogonal text anchors are constructed via linear projections, guiding cross-attention mechanisms to produce fused features. A hypernetwork further disentangles attribute-related information and generates instance-aware visual prompts, which encode dual-modal cues for fairness and robustness. Prototype-based regularization is applied in the visual branch to enforce separation of sensitive features and strengthen alignment with textual anchors. Extensive experiments on eight medical imaging datasets across four modalities show that DualFairVL achieves state-of-the-art fairness and accuracy under both in- and out-of-distribution settings, outperforming full fine-tuning and parameter-efficient baselines with only 3.6M trainable parameters. Code will be released upon publication.
Deformable Medical Image Registration with Effective Anatomical Structure Representation and Divide-and-Conquer Network
Jun 24, 2025Abstract:Effective representation of Regions of Interest (ROI) and independent alignment of these ROIs can significantly enhance the performance of deformable medical image registration (DMIR). However, current learning-based DMIR methods have limitations. Unsupervised techniques disregard ROI representation and proceed directly with aligning pairs of images, while weakly-supervised methods heavily depend on label constraints to facilitate registration. To address these issues, we introduce a novel ROI-based registration approach named EASR-DCN. Our method represents medical images through effective ROIs and achieves independent alignment of these ROIs without requiring labels. Specifically, we first used a Gaussian mixture model for intensity analysis to represent images using multiple effective ROIs with distinct intensities. Furthermore, we propose a novel Divide-and-Conquer Network (DCN) to process these ROIs through separate channels to learn feature alignments for each ROI. The resultant correspondences are seamlessly integrated to generate a comprehensive displacement vector field. Extensive experiments were performed on three MRI and one CT datasets to showcase the superior accuracy and deformation reduction efficacy of our EASR-DCN. Compared to VoxelMorph, our EASR-DCN achieved improvements of 10.31\% in the Dice score for brain MRI, 13.01\% for cardiac MRI, and 5.75\% for hippocampus MRI, highlighting its promising potential for clinical applications. The code for this work will be released upon acceptance of the paper.
Enjoying Information Dividend: Gaze Track-based Medical Weakly Supervised Segmentation
May 28, 2025Abstract:Weakly supervised semantic segmentation (WSSS) in medical imaging struggles with effectively using sparse annotations. One promising direction for WSSS leverages gaze annotations, captured via eye trackers that record regions of interest during diagnostic procedures. However, existing gaze-based methods, such as GazeMedSeg, do not fully exploit the rich information embedded in gaze data. In this paper, we propose GradTrack, a framework that utilizes physicians' gaze track, including fixation points, durations, and temporal order, to enhance WSSS performance. GradTrack comprises two key components: Gaze Track Map Generation and Track Attention, which collaboratively enable progressive feature refinement through multi-level gaze supervision during the decoding process. Experiments on the Kvasir-SEG and NCI-ISBI datasets demonstrate that GradTrack consistently outperforms existing gaze-based methods, achieving Dice score improvements of 3.21\% and 2.61\%, respectively. Moreover, GradTrack significantly narrows the performance gap with fully supervised models such as nnUNet.
SPP-SBL: Space-Power Prior Sparse Bayesian Learning for Block Sparse Recovery
May 13, 2025Abstract:The recovery of block-sparse signals with unknown structural patterns remains a fundamental challenge in structured sparse signal reconstruction. By proposing a variance transformation framework, this paper unifies existing pattern-based block sparse Bayesian learning methods, and introduces a novel space power prior based on undirected graph models to adaptively capture the unknown patterns of block-sparse signals. By combining the EM algorithm with high-order equation root-solving, we develop a new structured sparse Bayesian learning method, SPP-SBL, which effectively addresses the open problem of space coupling parameter estimation in pattern-based methods. We further demonstrate that learning the relative values of space coupling parameters is key to capturing unknown block-sparse patterns and improving recovery accuracy. Experiments validate that SPP-SBL successfully recovers various challenging structured sparse signals (e.g., chain-structured signals and multi-pattern sparse signals) and real-world multi-modal structured sparse signals (images, audio), showing significant advantages in recovery accuracy across multiple metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge