Shishuai Hu
Vision-Amplified Semantic Entropy for Hallucination Detection in Medical Visual Question Answering
Mar 26, 2025Abstract:Multimodal large language models (MLLMs) have demonstrated significant potential in medical Visual Question Answering (VQA). Yet, they remain prone to hallucinations-incorrect responses that contradict input images, posing substantial risks in clinical decision-making. Detecting these hallucinations is essential for establishing trust in MLLMs among clinicians and patients, thereby enabling their real-world adoption. Current hallucination detection methods, especially semantic entropy (SE), have demonstrated promising hallucination detection capacity for LLMs. However, adapting SE to medical MLLMs by incorporating visual perturbations presents a dilemma. Weak perturbations preserve image content and ensure clinical validity, but may be overlooked by medical MLLMs, which tend to over rely on language priors. In contrast, strong perturbations can distort essential diagnostic features, compromising clinical interpretation. To address this issue, we propose Vision Amplified Semantic Entropy (VASE), which incorporates weak image transformations and amplifies the impact of visual input, to improve hallucination detection in medical VQA. We first estimate the semantic predictive distribution under weak visual transformations to preserve clinical validity, and then amplify visual influence by contrasting this distribution with that derived from a distorted image. The entropy of the resulting distribution is estimated as VASE. Experiments on two medical open-ended VQA datasets demonstrate that VASE consistently outperforms existing hallucination detection methods.
Unleashing the Potential of Open-set Noisy Samples Against Label Noise for Medical Image Classification
Jun 18, 2024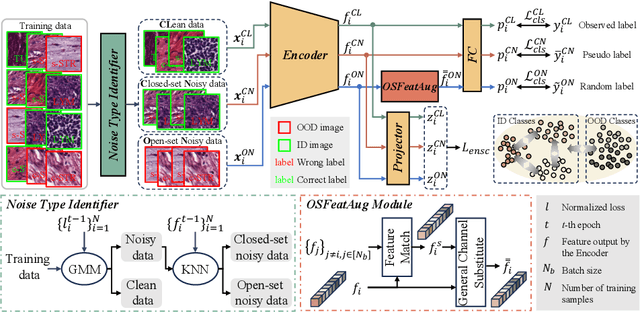
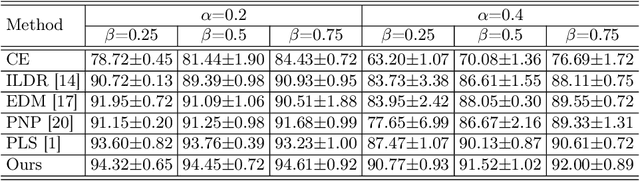
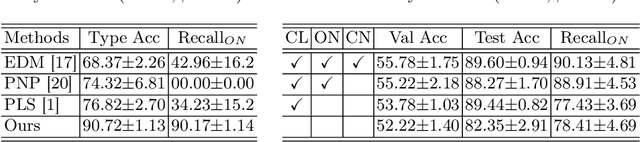
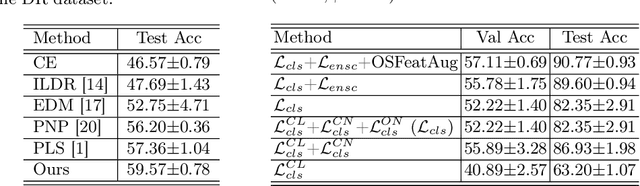
Abstract:The challenge of addressing mixed closed-set and open-set label noise in medical image classification remains largely unexplored. Unlike natural image classification where there is a common practice of segregation and separate processing of closed-set and open-set noisy samples from clean ones, medical image classification faces difficulties due to high inter-class similarity which complicates the identification of open-set noisy samples. Moreover, prevailing methods do not leverage the full potential of open-set noisy samples for label noise mitigation, often leading to their exclusion or application of uniform soft labels. To address these issues, we propose an Extended Noise-robust Contrastive and Open-set Feature Augmentation (ENCOFA) framework. ENCOFA includes the Extended Noise-robust Supervised Contrastive (ENSC) Loss, which aids in distinguishing features across classes. The ENSC loss regards open-set noisy samples as an extended class and mitigates label noise by weighting contrastive pairs with label reliability. Furthermore, we develop an Open-set Feature Augmentation (OSFeatAug) module that enriches the features of open-set samples, utilizing the model's extra capacity to prevent overfitting to noisy data. We conducted experiments on a synthetic noisy dataset and a real-world noisy dataset. Our results indicate the superiority of ENCOFA and the effectiveness of leveraging the open-set noisy samples to combat label noise.
Towards Clinician-Preferred Segmentation: Leveraging Human-in-the-Loop for Test Time Adaptation in Medical Image Segmentation
May 14, 2024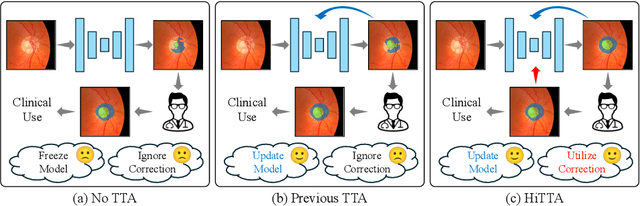
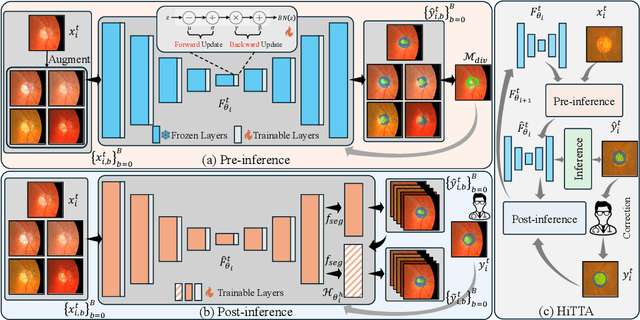
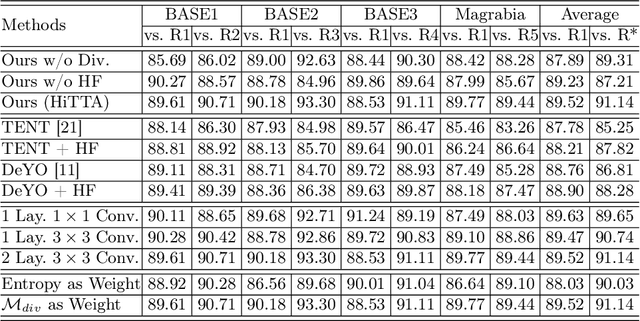
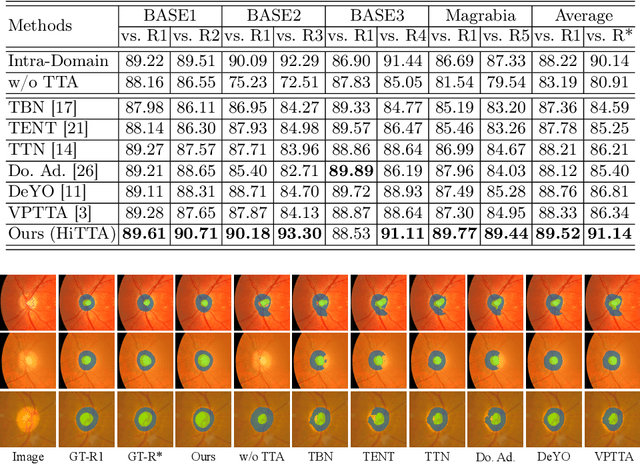
Abstract:Deep learning-based medical image segmentation models often face performance degradation when deployed across various medical centers, largely due to the discrepancies in data distribution. Test Time Adaptation (TTA) methods, which adapt pre-trained models to test data, have been employed to mitigate such discrepancies. However, existing TTA methods primarily focus on manipulating Batch Normalization (BN) layers or employing prompt and adversarial learning, which may not effectively rectify the inconsistencies arising from divergent data distributions. In this paper, we propose a novel Human-in-the-loop TTA (HiTTA) framework that stands out in two significant ways. First, it capitalizes on the largely overlooked potential of clinician-corrected predictions, integrating these corrections into the TTA process to steer the model towards predictions that coincide more closely with clinical annotation preferences. Second, our framework conceives a divergence loss, designed specifically to diminish the prediction divergence instigated by domain disparities, through the careful calibration of BN parameters. Our HiTTA is distinguished by its dual-faceted capability to acclimatize to the distribution of test data whilst ensuring the model's predictions align with clinical expectations, thereby enhancing its relevance in a medical context. Extensive experiments on a public dataset underscore the superiority of our HiTTA over existing TTA methods, emphasizing the advantages of integrating human feedback and our divergence loss in enhancing the model's performance and adaptability across diverse medical centers.
Devil is in Channels: Contrastive Single Domain Generalization for Medical Image Segmentation
Jun 24, 2023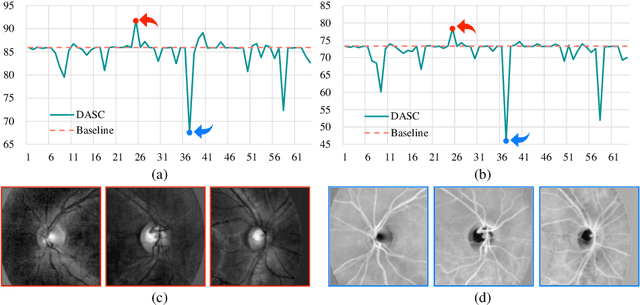
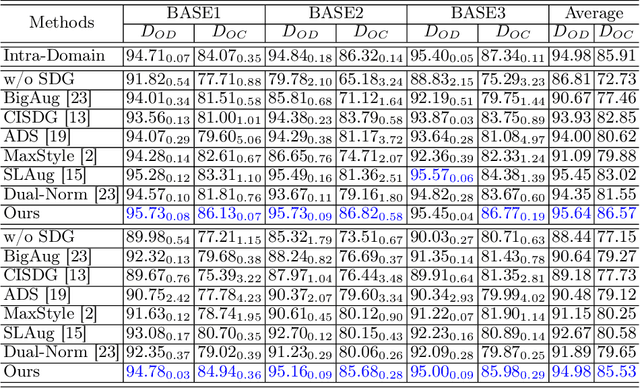
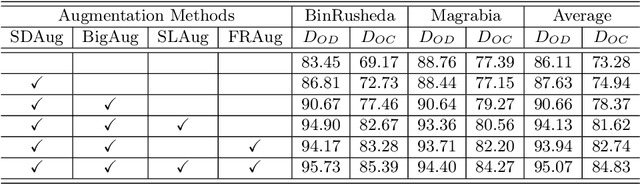
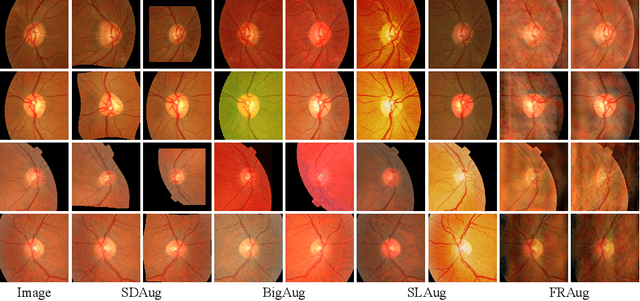
Abstract:Deep learning-based medical image segmentation models suffer from performance degradation when deployed to a new healthcare center. To address this issue, unsupervised domain adaptation and multi-source domain generalization methods have been proposed, which, however, are less favorable for clinical practice due to the cost of acquiring target-domain data and the privacy concerns associated with redistributing the data from multiple source domains. In this paper, we propose a \textbf{C}hannel-level \textbf{C}ontrastive \textbf{S}ingle \textbf{D}omain \textbf{G}eneralization (\textbf{C$^2$SDG}) model for medical image segmentation. In C$^2$SDG, the shallower features of each image and its style-augmented counterpart are extracted and used for contrastive training, resulting in the disentangled style representations and structure representations. The segmentation is performed based solely on the structure representations. Our method is novel in the contrastive perspective that enables channel-wise feature disentanglement using a single source domain. We evaluated C$^2$SDG against six SDG methods on a multi-domain joint optic cup and optic disc segmentation benchmark. Our results suggest the effectiveness of each module in C$^2$SDG and also indicate that C$^2$SDG outperforms the baseline and all competing methods with a large margin. The code will be available at \url{https://github.com/ShishuaiHu/CCSDG}.
Transformer-based Annotation Bias-aware Medical Image Segmentation
Jun 02, 2023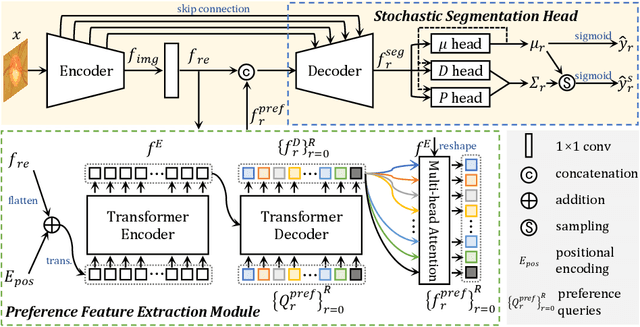
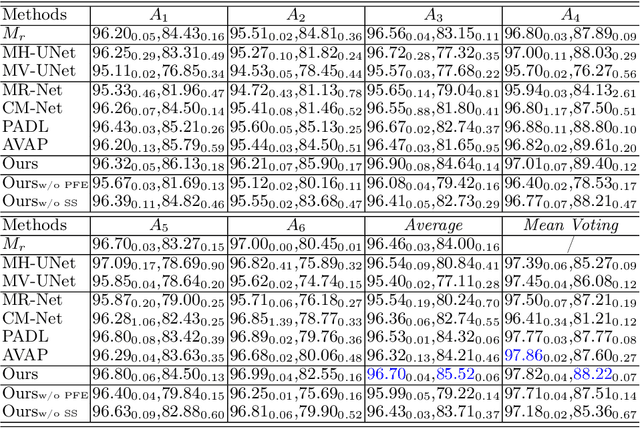
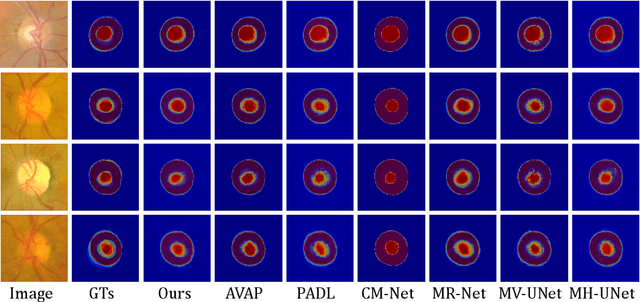
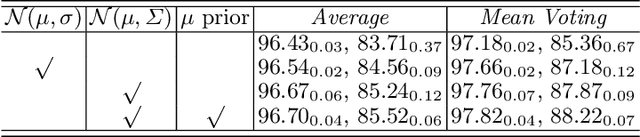
Abstract:Manual medical image segmentation is subjective and suffers from annotator-related bias, which can be mimicked or amplified by deep learning methods. Recently, researchers have suggested that such bias is the combination of the annotator preference and stochastic error, which are modeled by convolution blocks located after decoder and pixel-wise independent Gaussian distribution, respectively. It is unlikely that convolution blocks can effectively model the varying degrees of preference at the full resolution level. Additionally, the independent pixel-wise Gaussian distribution disregards pixel correlations, leading to a discontinuous boundary. This paper proposes a Transformer-based Annotation Bias-aware (TAB) medical image segmentation model, which tackles the annotator-related bias via modeling annotator preference and stochastic errors. TAB employs the Transformer with learnable queries to extract the different preference-focused features. This enables TAB to produce segmentation with various preferences simultaneously using a single segmentation head. Moreover, TAB takes the multivariant normal distribution assumption that models pixel correlations, and learns the annotation distribution to disentangle the stochastic error. We evaluated our TAB on an OD/OC segmentation benchmark annotated by six annotators. Our results suggest that TAB outperforms existing medical image segmentation models which take into account the annotator-related bias.
Instance-specific Label Distribution Regularization for Learning with Label Noise
Dec 16, 2022



Abstract:Modeling noise transition matrix is a kind of promising method for learning with label noise. Based on the estimated noise transition matrix and the noisy posterior probabilities, the clean posterior probabilities, which are jointly called Label Distribution (LD) in this paper, can be calculated as the supervision. To reliably estimate the noise transition matrix, some methods assume that anchor points are available during training. Nonetheless, if anchor points are invalid, the noise transition matrix might be poorly learned, resulting in poor performance. Consequently, other methods treat reliable data points, extracted from training data, as pseudo anchor points. However, from a statistical point of view, the noise transition matrix can be inferred from data with noisy labels under the clean-label-domination assumption. Therefore, we aim to estimate the noise transition matrix without (pseudo) anchor points. There is evidence showing that samples are more likely to be mislabeled as other similar class labels, which means the mislabeling probability is highly correlated with the inter-class correlation. Inspired by this observation, we propose an instance-specific Label Distribution Regularization (LDR), in which the instance-specific LD is estimated as the supervision, to prevent DCNNs from memorizing noisy labels. Specifically, we estimate the noisy posterior under the supervision of noisy labels, and approximate the batch-level noise transition matrix by estimating the inter-class correlation matrix with neither anchor points nor pseudo anchor points. Experimental results on two synthetic noisy datasets and two real-world noisy datasets demonstrate that our LDR outperforms existing methods.
ProSFDA: Prompt Learning based Source-free Domain Adaptation for Medical Image Segmentation
Nov 21, 2022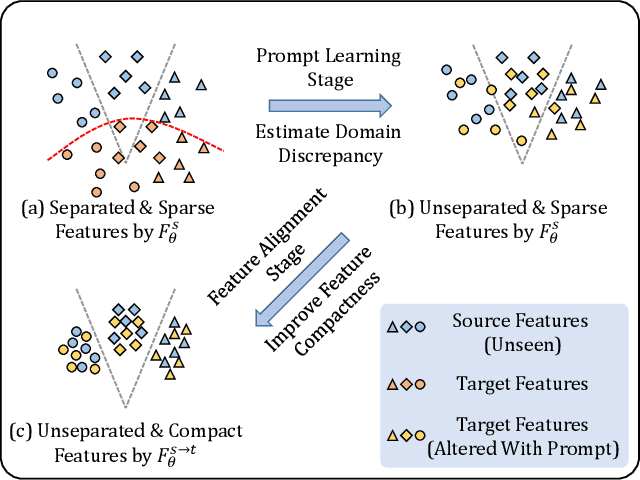
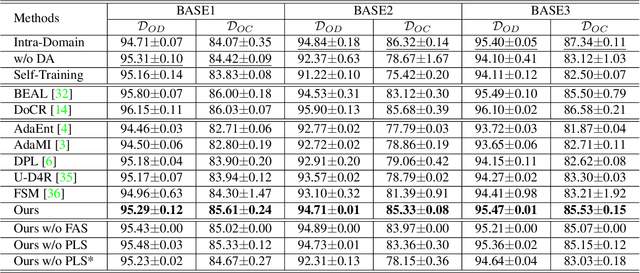
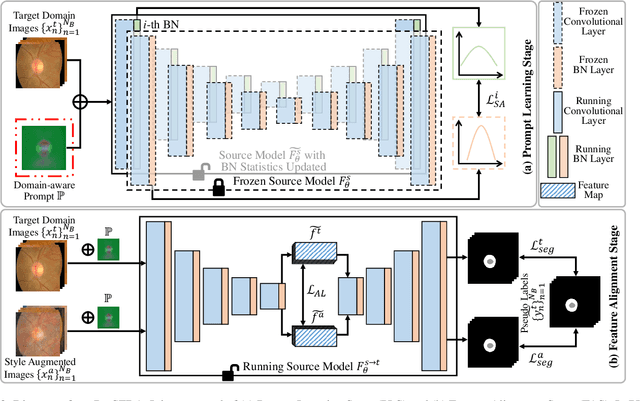
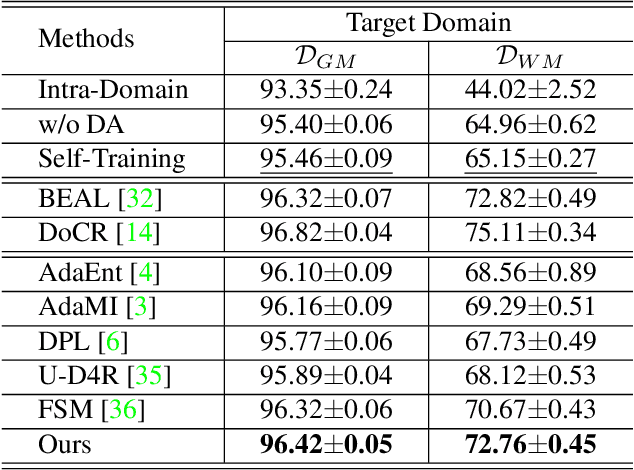
Abstract:The domain discrepancy existed between medical images acquired in different situations renders a major hurdle in deploying pre-trained medical image segmentation models for clinical use. Since it is less possible to distribute training data with the pre-trained model due to the huge data size and privacy concern, source-free unsupervised domain adaptation (SFDA) has recently been increasingly studied based on either pseudo labels or prior knowledge. However, the image features and probability maps used by pseudo label-based SFDA and the consistent prior assumption and the prior prediction network used by prior-guided SFDA may become less reliable when the domain discrepancy is large. In this paper, we propose a \textbf{Pro}mpt learning based \textbf{SFDA} (\textbf{ProSFDA}) method for medical image segmentation, which aims to improve the quality of domain adaption by minimizing explicitly the domain discrepancy. Specifically, in the prompt learning stage, we estimate source-domain images via adding a domain-aware prompt to target-domain images, then optimize the prompt via minimizing the statistic alignment loss, and thereby prompt the source model to generate reliable predictions on (altered) target-domain images. In the feature alignment stage, we also align the features of target-domain images and their styles-augmented counterparts to optimize the source model, and hence push the model to extract compact features. We evaluate our ProSFDA on two multi-domain medical image segmentation benchmarks. Our results indicate that the proposed ProSFDA outperforms substantially other SFDA methods and is even comparable to UDA methods. Code will be available at \url{https://github.com/ShishuaiHu/ProSFDA}.
Boundary-Aware Network for Abdominal Multi-Organ Segmentation
Aug 29, 2022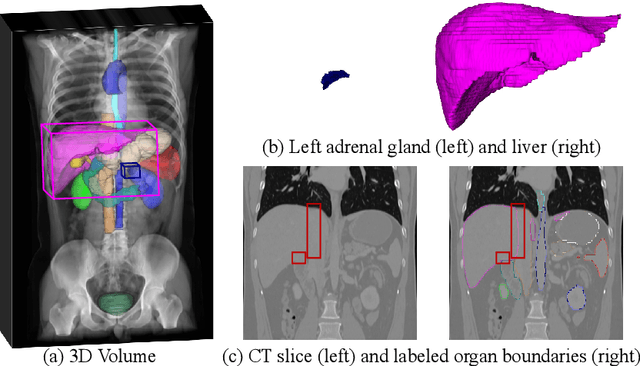
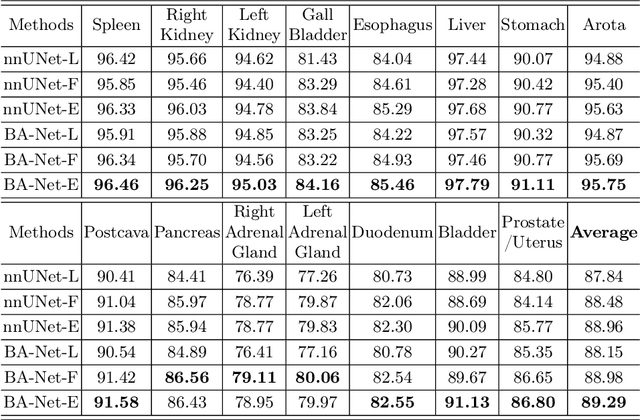
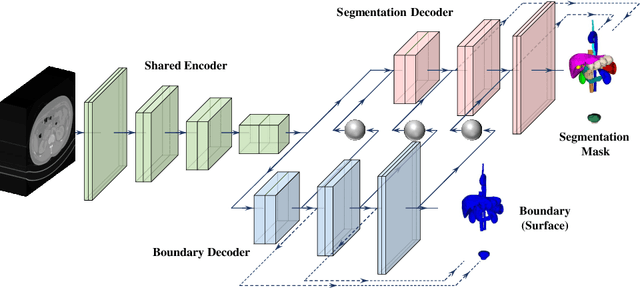
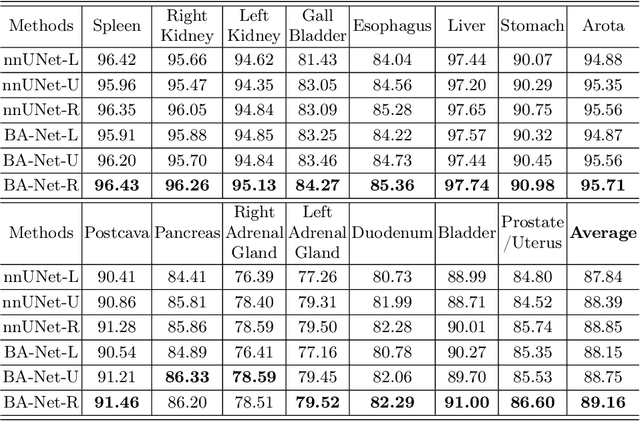
Abstract:Automated abdominal multi-organ segmentation is a crucial yet challenging task in the computer-aided diagnosis of abdominal organ-related diseases. Although numerous deep learning models have achieved remarkable success in many medical image segmentation tasks, accurate segmentation of abdominal organs remains challenging, due to the varying sizes of abdominal organs and the ambiguous boundaries among them. In this paper, we propose a boundary-aware network (BA-Net) to segment abdominal organs on CT scans and MRI scans. This model contains a shared encoder, a boundary decoder, and a segmentation decoder. The multi-scale deep supervision strategy is adopted on both decoders, which can alleviate the issues caused by variable organ sizes. The boundary probability maps produced by the boundary decoder at each scale are used as attention to enhance the segmentation feature maps. We evaluated the BA-Net on the Abdominal Multi-Organ Segmentation (AMOS) Challenge dataset and achieved an average Dice score of 89.29$\%$ for multi-organ segmentation on CT scans and an average Dice score of 71.92$\%$ on MRI scans. The results demonstrate that BA-Net is superior to nnUNet on both segmentation tasks.
Boundary-Aware Network for Kidney Parsing
Aug 29, 2022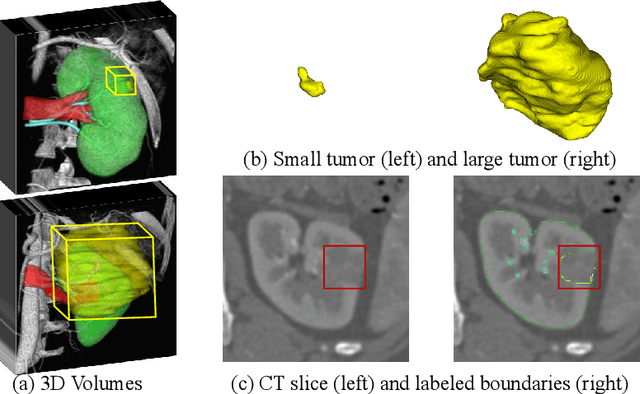
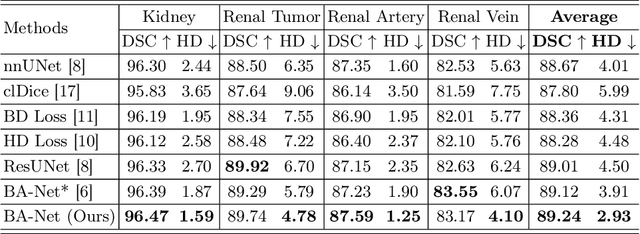
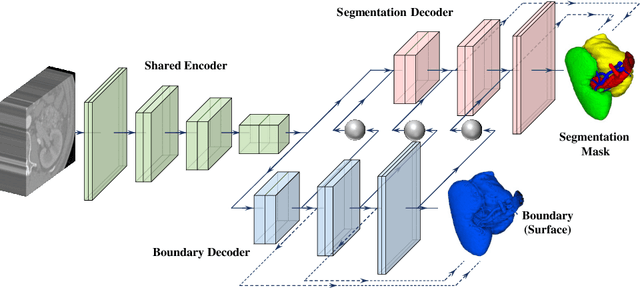
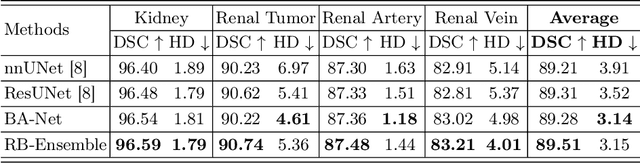
Abstract:Kidney structures segmentation is a crucial yet challenging task in the computer-aided diagnosis of surgery-based renal cancer. Although numerous deep learning models have achieved remarkable success in many medical image segmentation tasks, accurate segmentation of kidney structures on computed tomography angiography (CTA) images remains challenging, due to the variable sizes of kidney tumors and the ambiguous boundaries between kidney structures and their surroundings. In this paper, we propose a boundary-aware network (BA-Net) to segment kidneys, kidney tumors, arteries, and veins on CTA scans. This model contains a shared encoder, a boundary decoder, and a segmentation decoder. The multi-scale deep supervision strategy is adopted on both decoders, which can alleviate the issues caused by variable tumor sizes. The boundary probability maps produced by the boundary decoder at each scale are used as attention to enhance the segmentation feature maps. We evaluated the BA-Net on the Kidney PArsing (KiPA) Challenge dataset and achieved an average Dice score of 89.65$\%$ for kidney structure segmentation on CTA scans using 4-fold cross-validation. The results demonstrate the effectiveness of the BA-Net.
Label Propagation for 3D Carotid Vessel Wall Segmentation and Atherosclerosis Diagnosis
Aug 29, 2022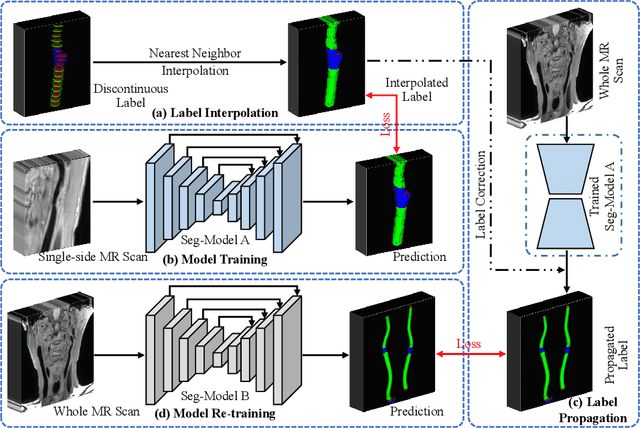
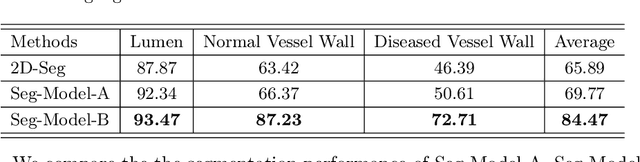
Abstract:Carotid vessel wall segmentation is a crucial yet challenging task in the computer-aided diagnosis of atherosclerosis. Although numerous deep learning models have achieved remarkable success in many medical image segmentation tasks, accurate segmentation of carotid vessel wall on magnetic resonance (MR) images remains challenging, due to limited annotations and heterogeneous arteries. In this paper, we propose a semi-supervised label propagation framework to segment lumen, normal vessel walls, and atherosclerotic vessel wall on 3D MR images. By interpolating the provided annotations, we get 3D continuous labels for training 3D segmentation model. With the trained model, we generate pseudo labels for unlabeled slices to incorporate them for model training. Then we use the whole MR scans and the propagated labels to re-train the segmentation model and improve its robustness. We evaluated the label propagation framework on the CarOtid vessel wall SegMentation and atherosclerOsis diagnosiS (COSMOS) Challenge dataset and achieved a QuanM score of 83.41\% on the testing dataset, which got the 1-st place on the online evaluation leaderboard. The results demonstrate the effectiveness of the proposed framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge