Huazhu Fu
Structure-constrained Language-informed Diffusion Model for Unpaired Low-dose Computed Tomography Angiography Reconstruction
Jan 28, 2026Abstract:The application of iodinated contrast media (ICM) improves the sensitivity and specificity of computed tomography (CT) for a wide range of clinical indications. However, overdose of ICM can cause problems such as kidney damage and life-threatening allergic reactions. Deep learning methods can generate CT images of normal-dose ICM from low-dose ICM, reducing the required dose while maintaining diagnostic power. However, existing methods are difficult to realize accurate enhancement with incompletely paired images, mainly because of the limited ability of the model to recognize specific structures. To overcome this limitation, we propose a Structure-constrained Language-informed Diffusion Model (SLDM), a unified medical generation model that integrates structural synergy and spatial intelligence. First, the structural prior information of the image is effectively extracted to constrain the model inference process, thus ensuring structural consistency in the enhancement process. Subsequently, semantic supervision strategy with spatial intelligence is introduced, which integrates the functions of visual perception and spatial reasoning, thus prompting the model to achieve accurate enhancement. Finally, the subtraction angiography enhancement module is applied, which serves to improve the contrast of the ICM agent region to suitable interval for observation. Qualitative analysis of visual comparison and quantitative results of several metrics demonstrate the effectiveness of our method in angiographic reconstruction for low-dose contrast medium CT angiography.
Topology-aware Pathological Consistency Matching for Weakly-Paired IHC Virtual Staining
Jan 06, 2026Abstract:Immunohistochemical (IHC) staining provides crucial molecular characterization of tissue samples and plays an indispensable role in the clinical examination and diagnosis of cancers. However, compared with the commonly used Hematoxylin and Eosin (H&E) staining, IHC staining involves complex procedures and is both time-consuming and expensive, which limits its widespread clinical use. Virtual staining converts H&E images to IHC images, offering a cost-effective alternative to clinical IHC staining. Nevertheless, using adjacent slides as ground truth often results in weakly-paired data with spatial misalignment and local deformations, hindering effective supervised learning. To address these challenges, we propose a novel topology-aware framework for H&E-to-IHC virtual staining. Specifically, we introduce a Topology-aware Consistency Matching (TACM) mechanism that employs graph contrastive learning and topological perturbations to learn robust matching patterns despite spatial misalignments, ensuring structural consistency. Furthermore, we propose a Topology-constrained Pathological Matching (TCPM) mechanism that aligns pathological positive regions based on node importance to enhance pathological consistency. Extensive experiments on two benchmarks across four staining tasks demonstrate that our method outperforms state-of-the-art approaches, achieving superior generation quality with higher clinical relevance.
Plasticine: A Traceable Diffusion Model for Medical Image Translation
Dec 20, 2025Abstract:Domain gaps arising from variations in imaging devices and population distributions pose significant challenges for machine learning in medical image analysis. Existing image-to-image translation methods primarily aim to learn mappings between domains, often generating diverse synthetic data with variations in anatomical scale and shape, but they usually overlook spatial correspondence during the translation process. For clinical applications, traceability, defined as the ability to provide pixel-level correspondences between original and translated images, is equally important. This property enhances clinical interpretability but has been largely overlooked in previous approaches. To address this gap, we propose Plasticine, which is, to the best of our knowledge, the first end-to-end image-to-image translation framework explicitly designed with traceability as a core objective. Our method combines intensity translation and spatial transformation within a denoising diffusion framework. This design enables the generation of synthetic images with interpretable intensity transitions and spatially coherent deformations, supporting pixel-wise traceability throughout the translation process.
Probing Scientific General Intelligence of LLMs with Scientist-Aligned Workflows
Dec 18, 2025Abstract:Despite advances in scientific AI, a coherent framework for Scientific General Intelligence (SGI)-the ability to autonomously conceive, investigate, and reason across scientific domains-remains lacking. We present an operational SGI definition grounded in the Practical Inquiry Model (PIM: Deliberation, Conception, Action, Perception) and operationalize it via four scientist-aligned tasks: deep research, idea generation, dry/wet experiments, and experimental reasoning. SGI-Bench comprises over 1,000 expert-curated, cross-disciplinary samples inspired by Science's 125 Big Questions, enabling systematic evaluation of state-of-the-art LLMs. Results reveal gaps: low exact match (10--20%) in deep research despite step-level alignment; ideas lacking feasibility and detail; high code executability but low execution result accuracy in dry experiments; low sequence fidelity in wet protocols; and persistent multimodal comparative-reasoning challenges. We further introduce Test-Time Reinforcement Learning (TTRL), which optimizes retrieval-augmented novelty rewards at inference, enhancing hypothesis novelty without reference answer. Together, our PIM-grounded definition, workflow-centric benchmark, and empirical insights establish a foundation for AI systems that genuinely participate in scientific discovery.
Improving Learning of New Diseases through Knowledge-Enhanced Initialization for Federated Adapter Tuning
Aug 14, 2025

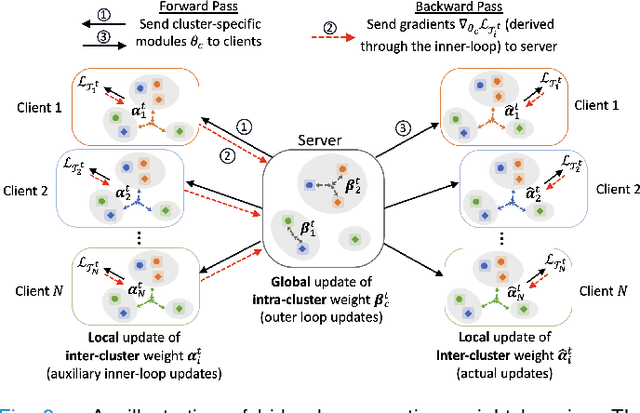
Abstract:In healthcare, federated learning (FL) is a widely adopted framework that enables privacy-preserving collaboration among medical institutions. With large foundation models (FMs) demonstrating impressive capabilities, using FMs in FL through cost-efficient adapter tuning has become a popular approach. Given the rapidly evolving healthcare environment, it is crucial for individual clients to quickly adapt to new tasks or diseases by tuning adapters while drawing upon past experiences. In this work, we introduce Federated Knowledge-Enhanced Initialization (FedKEI), a novel framework that leverages cross-client and cross-task transfer from past knowledge to generate informed initializations for learning new tasks with adapters. FedKEI begins with a global clustering process at the server to generalize knowledge across tasks, followed by the optimization of aggregation weights across clusters (inter-cluster weights) and within each cluster (intra-cluster weights) to personalize knowledge transfer for each new task. To facilitate more effective learning of the inter- and intra-cluster weights, we adopt a bi-level optimization scheme that collaboratively learns the global intra-cluster weights across clients and optimizes the local inter-cluster weights toward each client's task objective. Extensive experiments on three benchmark datasets of different modalities, including dermatology, chest X-rays, and retinal OCT, demonstrate FedKEI's advantage in adapting to new diseases compared to state-of-the-art methods.
Uncertainty-aware Cross-training for Semi-supervised Medical Image Segmentation
Aug 12, 2025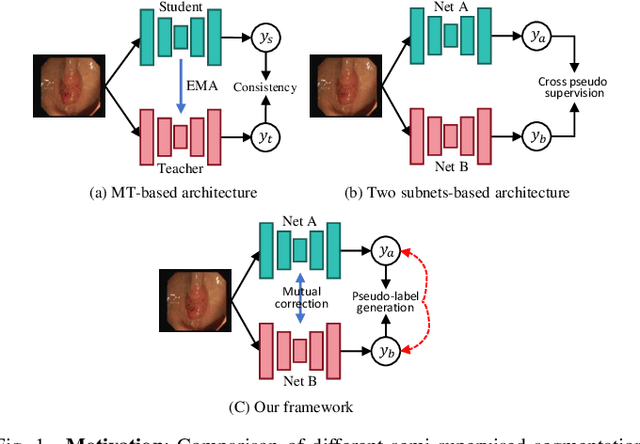
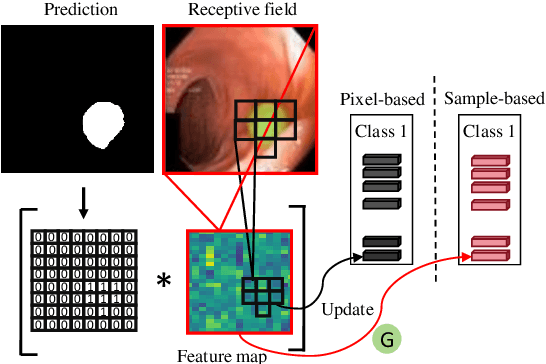
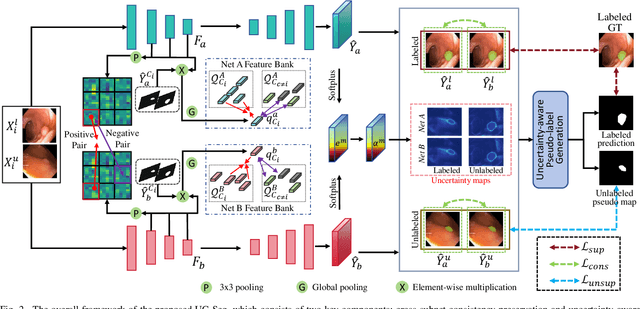
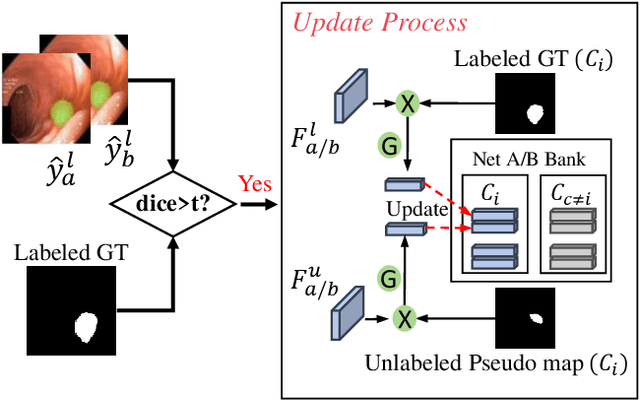
Abstract:Semi-supervised learning has gained considerable popularity in medical image segmentation tasks due to its capability to reduce reliance on expert-examined annotations. Several mean-teacher (MT) based semi-supervised methods utilize consistency regularization to effectively leverage valuable information from unlabeled data. However, these methods often heavily rely on the student model and overlook the potential impact of cognitive biases within the model. Furthermore, some methods employ co-training using pseudo-labels derived from different inputs, yet generating high-confidence pseudo-labels from perturbed inputs during training remains a significant challenge. In this paper, we propose an Uncertainty-aware Cross-training framework for semi-supervised medical image Segmentation (UC-Seg). Our UC-Seg framework incorporates two distinct subnets to effectively explore and leverage the correlation between them, thereby mitigating cognitive biases within the model. Specifically, we present a Cross-subnet Consistency Preservation (CCP) strategy to enhance feature representation capability and ensure feature consistency across the two subnets. This strategy enables each subnet to correct its own biases and learn shared semantics from both labeled and unlabeled data. Additionally, we propose an Uncertainty-aware Pseudo-label Generation (UPG) component that leverages segmentation results and corresponding uncertainty maps from both subnets to generate high-confidence pseudo-labels. We extensively evaluate the proposed UC-Seg on various medical image segmentation tasks involving different modality images, such as MRI, CT, ultrasound, colonoscopy, and so on. The results demonstrate that our method achieves superior segmentation accuracy and generalization performance compared to other state-of-the-art semi-supervised methods. Our code will be released at https://github.com/taozh2017/UCSeg.
AdaFusion: Prompt-Guided Inference with Adaptive Fusion of Pathology Foundation Models
Aug 07, 2025Abstract:Pathology foundation models (PFMs) have demonstrated strong representational capabilities through self-supervised pre-training on large-scale, unannotated histopathology image datasets. However, their diverse yet opaque pretraining contexts, shaped by both data-related and structural/training factors, introduce latent biases that hinder generalisability and transparency in downstream applications. In this paper, we propose AdaFusion, a novel prompt-guided inference framework that, to our knowledge, is among the very first to dynamically integrate complementary knowledge from multiple PFMs. Our method compresses and aligns tile-level features from diverse models and employs a lightweight attention mechanism to adaptively fuse them based on tissue phenotype context. We evaluate AdaFusion on three real-world benchmarks spanning treatment response prediction, tumour grading, and spatial gene expression inference. Our approach consistently surpasses individual PFMs across both classification and regression tasks, while offering interpretable insights into each model's biosemantic specialisation. These results highlight AdaFusion's ability to bridge heterogeneous PFMs, achieving both enhanced performance and interpretability of model-specific inductive biases.
Text-driven Multiplanar Visual Interaction for Semi-supervised Medical Image Segmentation
Jul 16, 2025Abstract:Semi-supervised medical image segmentation is a crucial technique for alleviating the high cost of data annotation. When labeled data is limited, textual information can provide additional context to enhance visual semantic understanding. However, research exploring the use of textual data to enhance visual semantic embeddings in 3D medical imaging tasks remains scarce. In this paper, we propose a novel text-driven multiplanar visual interaction framework for semi-supervised medical image segmentation (termed Text-SemiSeg), which consists of three main modules: Text-enhanced Multiplanar Representation (TMR), Category-aware Semantic Alignment (CSA), and Dynamic Cognitive Augmentation (DCA). Specifically, TMR facilitates text-visual interaction through planar mapping, thereby enhancing the category awareness of visual features. CSA performs cross-modal semantic alignment between the text features with introduced learnable variables and the intermediate layer of visual features. DCA reduces the distribution discrepancy between labeled and unlabeled data through their interaction, thus improving the model's robustness. Finally, experiments on three public datasets demonstrate that our model effectively enhances visual features with textual information and outperforms other methods. Our code is available at https://github.com/taozh2017/Text-SemiSeg.
APTOS-2024 challenge report: Generation of synthetic 3D OCT images from fundus photographs
Jun 09, 2025Abstract:Optical Coherence Tomography (OCT) provides high-resolution, 3D, and non-invasive visualization of retinal layers in vivo, serving as a critical tool for lesion localization and disease diagnosis. However, its widespread adoption is limited by equipment costs and the need for specialized operators. In comparison, 2D color fundus photography offers faster acquisition and greater accessibility with less dependence on expensive devices. Although generative artificial intelligence has demonstrated promising results in medical image synthesis, translating 2D fundus images into 3D OCT images presents unique challenges due to inherent differences in data dimensionality and biological information between modalities. To advance generative models in the fundus-to-3D-OCT setting, the Asia Pacific Tele-Ophthalmology Society (APTOS-2024) organized a challenge titled Artificial Intelligence-based OCT Generation from Fundus Images. This paper details the challenge framework (referred to as APTOS-2024 Challenge), including: the benchmark dataset, evaluation methodology featuring two fidelity metrics-image-based distance (pixel-level OCT B-scan similarity) and video-based distance (semantic-level volumetric consistency), and analysis of top-performing solutions. The challenge attracted 342 participating teams, with 42 preliminary submissions and 9 finalists. Leading methodologies incorporated innovations in hybrid data preprocessing or augmentation (cross-modality collaborative paradigms), pre-training on external ophthalmic imaging datasets, integration of vision foundation models, and model architecture improvement. The APTOS-2024 Challenge is the first benchmark demonstrating the feasibility of fundus-to-3D-OCT synthesis as a potential solution for improving ophthalmic care accessibility in under-resourced healthcare settings, while helping to expedite medical research and clinical applications.
PathFL: Multi-Alignment Federated Learning for Pathology Image Segmentation
May 28, 2025Abstract:Pathology image segmentation across multiple centers encounters significant challenges due to diverse sources of heterogeneity including imaging modalities, organs, and scanning equipment, whose variability brings representation bias and impedes the development of generalizable segmentation models. In this paper, we propose PathFL, a novel multi-alignment Federated Learning framework for pathology image segmentation that addresses these challenges through three-level alignment strategies of image, feature, and model aggregation. Firstly, at the image level, a collaborative style enhancement module aligns and diversifies local data by facilitating style information exchange across clients. Secondly, at the feature level, an adaptive feature alignment module ensures implicit alignment in the representation space by infusing local features with global insights, promoting consistency across heterogeneous client features learning. Finally, at the model aggregation level, a stratified similarity aggregation strategy hierarchically aligns and aggregates models on the server, using layer-specific similarity to account for client discrepancies and enhance global generalization. Comprehensive evaluations on four sets of heterogeneous pathology image datasets, encompassing cross-source, cross-modality, cross-organ, and cross-scanner variations, validate the effectiveness of our PathFL in achieving better performance and robustness against data heterogeneity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge