Yueming Jin
SurgWorld: Learning Surgical Robot Policies from Videos via World Modeling
Dec 30, 2025Abstract:Data scarcity remains a fundamental barrier to achieving fully autonomous surgical robots. While large scale vision language action (VLA) models have shown impressive generalization in household and industrial manipulation by leveraging paired video action data from diverse domains, surgical robotics suffers from the paucity of datasets that include both visual observations and accurate robot kinematics. In contrast, vast corpora of surgical videos exist, but they lack corresponding action labels, preventing direct application of imitation learning or VLA training. In this work, we aim to alleviate this problem by learning policy models from SurgWorld, a world model designed for surgical physical AI. We curated the Surgical Action Text Alignment (SATA) dataset with detailed action description specifically for surgical robots. Then we built SurgeWorld based on the most advanced physical AI world model and SATA. It's able to generate diverse, generalizable and realistic surgery videos. We are also the first to use an inverse dynamics model to infer pseudokinematics from synthetic surgical videos, producing synthetic paired video action data. We demonstrate that a surgical VLA policy trained with these augmented data significantly outperforms models trained only on real demonstrations on a real surgical robot platform. Our approach offers a scalable path toward autonomous surgical skill acquisition by leveraging the abundance of unlabeled surgical video and generative world modeling, thus opening the door to generalizable and data efficient surgical robot policies.
Unleashing the Power of Image-Tabular Self-Supervised Learning via Breaking Cross-Tabular Barriers
Dec 16, 2025Abstract:Multi-modal learning integrating medical images and tabular data has significantly advanced clinical decision-making in recent years. Self-Supervised Learning (SSL) has emerged as a powerful paradigm for pretraining these models on large-scale unlabeled image-tabular data, aiming to learn discriminative representations. However, existing SSL methods for image-tabular representation learning are often confined to specific data cohorts, mainly due to their rigid tabular modeling mechanisms when modeling heterogeneous tabular data. This inter-tabular barrier hinders the multi-modal SSL methods from effectively learning transferrable medical knowledge shared across diverse cohorts. In this paper, we propose a novel SSL framework, namely CITab, designed to learn powerful multi-modal feature representations in a cross-tabular manner. We design the tabular modeling mechanism from a semantic-awareness perspective by integrating column headers as semantic cues, which facilitates transferrable knowledge learning and the scalability in utilizing multiple data sources for pretraining. Additionally, we propose a prototype-guided mixture-of-linear layer (P-MoLin) module for tabular feature specialization, empowering the model to effectively handle the heterogeneity of tabular data and explore the underlying medical concepts. We conduct comprehensive evaluations on Alzheimer's disease diagnosis task across three publicly available data cohorts containing 4,461 subjects. Experimental results demonstrate that CITab outperforms state-of-the-art approaches, paving the way for effective and scalable cross-tabular multi-modal learning.
Scaling Up Occupancy-centric Driving Scene Generation: Dataset and Method
Oct 27, 2025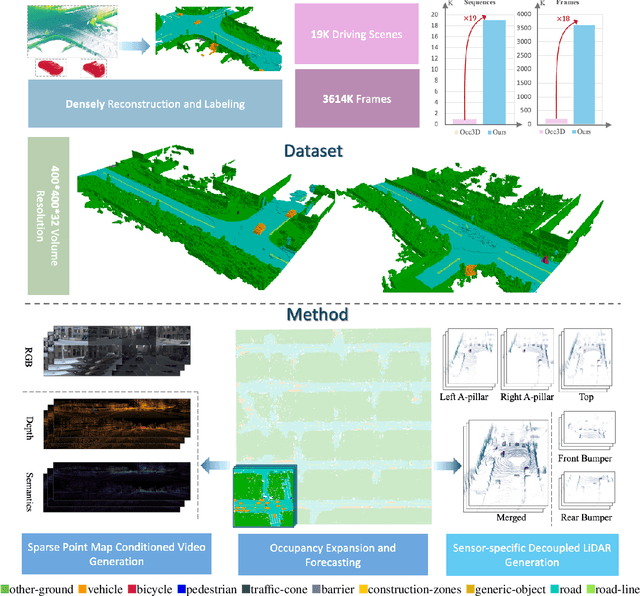
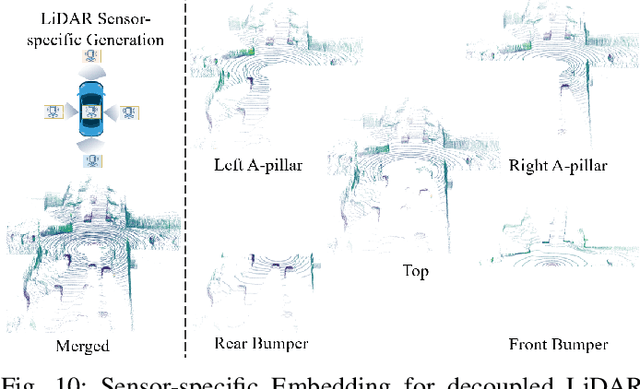
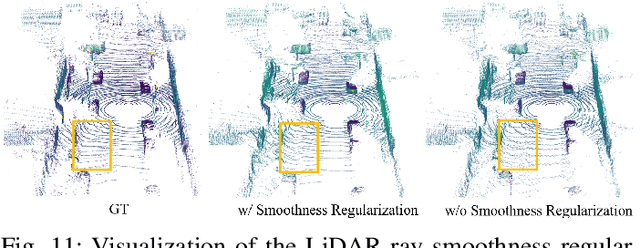
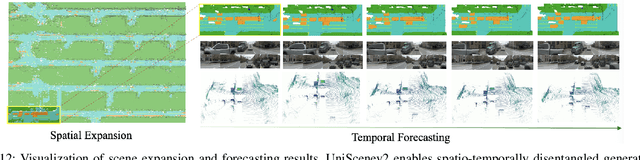
Abstract:Driving scene generation is a critical domain for autonomous driving, enabling downstream applications, including perception and planning evaluation. Occupancy-centric methods have recently achieved state-of-the-art results by offering consistent conditioning across frames and modalities; however, their performance heavily depends on annotated occupancy data, which still remains scarce. To overcome this limitation, we curate Nuplan-Occ, the largest semantic occupancy dataset to date, constructed from the widely used Nuplan benchmark. Its scale and diversity facilitate not only large-scale generative modeling but also autonomous driving downstream applications. Based on this dataset, we develop a unified framework that jointly synthesizes high-quality semantic occupancy, multi-view videos, and LiDAR point clouds. Our approach incorporates a spatio-temporal disentangled architecture to support high-fidelity spatial expansion and temporal forecasting of 4D dynamic occupancy. To bridge modal gaps, we further propose two novel techniques: a Gaussian splatting-based sparse point map rendering strategy that enhances multi-view video generation, and a sensor-aware embedding strategy that explicitly models LiDAR sensor properties for realistic multi-LiDAR simulation. Extensive experiments demonstrate that our method achieves superior generation fidelity and scalability compared to existing approaches, and validates its practical value in downstream tasks. Repo: https://github.com/Arlo0o/UniScene-Unified-Occupancy-centric-Driving-Scene-Generation/tree/v2
Generalized Deep Multi-view Clustering via Causal Learning with Partially Aligned Cross-view Correspondence
Sep 19, 2025Abstract:Multi-view clustering (MVC) aims to explore the common clustering structure across multiple views. Many existing MVC methods heavily rely on the assumption of view consistency, where alignments for corresponding samples across different views are ordered in advance. However, real-world scenarios often present a challenge as only partial data is consistently aligned across different views, restricting the overall clustering performance. In this work, we consider the model performance decreasing phenomenon caused by data order shift (i.e., from fully to partially aligned) as a generalized multi-view clustering problem. To tackle this problem, we design a causal multi-view clustering network, termed CauMVC. We adopt a causal modeling approach to understand multi-view clustering procedure. To be specific, we formulate the partially aligned data as an intervention and multi-view clustering with partially aligned data as an post-intervention inference. However, obtaining invariant features directly can be challenging. Thus, we design a Variational Auto-Encoder for causal learning by incorporating an encoder from existing information to estimate the invariant features. Moreover, a decoder is designed to perform the post-intervention inference. Lastly, we design a contrastive regularizer to capture sample correlations. To the best of our knowledge, this paper is the first work to deal generalized multi-view clustering via causal learning. Empirical experiments on both fully and partially aligned data illustrate the strong generalization and effectiveness of CauMVC.
BCRNet: Enhancing Landmark Detection in Laparoscopic Liver Surgery via Bezier Curve Refinement
Jun 18, 2025


Abstract:Laparoscopic liver surgery, while minimally invasive, poses significant challenges in accurately identifying critical anatomical structures. Augmented reality (AR) systems, integrating MRI/CT with laparoscopic images based on 2D-3D registration, offer a promising solution for enhancing surgical navigation. A vital aspect of the registration progress is the precise detection of curvilinear anatomical landmarks in laparoscopic images. In this paper, we propose BCRNet (Bezier Curve Refinement Net), a novel framework that significantly enhances landmark detection in laparoscopic liver surgery primarily via the Bezier curve refinement strategy. The framework starts with a Multi-modal Feature Extraction (MFE) module designed to robustly capture semantic features. Then we propose Adaptive Curve Proposal Initialization (ACPI) to generate pixel-aligned Bezier curves and confidence scores for reliable initial proposals. Additionally, we design the Hierarchical Curve Refinement (HCR) mechanism to enhance these proposals iteratively through a multi-stage process, capturing fine-grained contextual details from multi-scale pixel-level features for precise Bezier curve adjustment. Extensive evaluations on the L3D and P2ILF datasets demonstrate that BCRNet outperforms state-of-the-art methods, achieving significant performance improvements. Code will be available.
Automatically Identify and Rectify: Robust Deep Contrastive Multi-view Clustering in Noisy Scenarios
May 27, 2025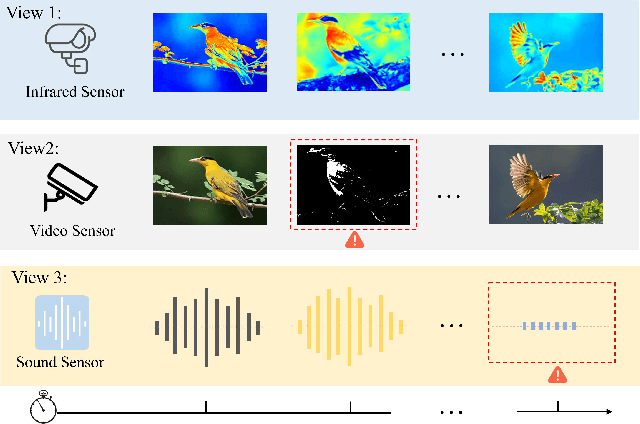
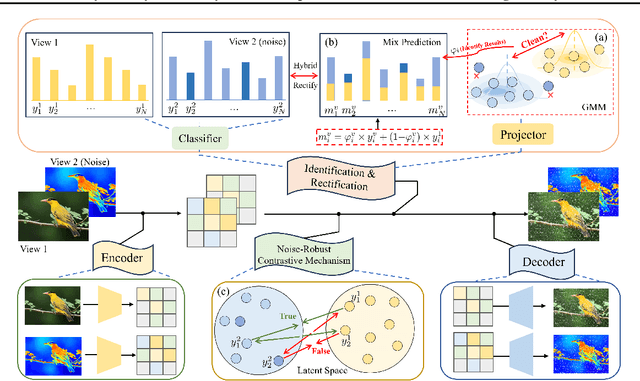

Abstract:Leveraging the powerful representation learning capabilities, deep multi-view clustering methods have demonstrated reliable performance by effectively integrating multi-source information from diverse views in recent years. Most existing methods rely on the assumption of clean views. However, noise is pervasive in real-world scenarios, leading to a significant degradation in performance. To tackle this problem, we propose a novel multi-view clustering framework for the automatic identification and rectification of noisy data, termed AIRMVC. Specifically, we reformulate noisy identification as an anomaly identification problem using GMM. We then design a hybrid rectification strategy to mitigate the adverse effects of noisy data based on the identification results. Furthermore, we introduce a noise-robust contrastive mechanism to generate reliable representations. Additionally, we provide a theoretical proof demonstrating that these representations can discard noisy information, thereby improving the performance of downstream tasks. Extensive experiments on six benchmark datasets demonstrate that AIRMVC outperforms state-of-the-art algorithms in terms of robustness in noisy scenarios. The code of AIRMVC are available at https://github.com/xihongyang1999/AIRMVC on Github.
ReSurgSAM2: Referring Segment Anything in Surgical Video via Credible Long-term Tracking
May 13, 2025Abstract:Surgical scene segmentation is critical in computer-assisted surgery and is vital for enhancing surgical quality and patient outcomes. Recently, referring surgical segmentation is emerging, given its advantage of providing surgeons with an interactive experience to segment the target object. However, existing methods are limited by low efficiency and short-term tracking, hindering their applicability in complex real-world surgical scenarios. In this paper, we introduce ReSurgSAM2, a two-stage surgical referring segmentation framework that leverages Segment Anything Model 2 to perform text-referred target detection, followed by tracking with reliable initial frame identification and diversity-driven long-term memory. For the detection stage, we propose a cross-modal spatial-temporal Mamba to generate precise detection and segmentation results. Based on these results, our credible initial frame selection strategy identifies the reliable frame for the subsequent tracking. Upon selecting the initial frame, our method transitions to the tracking stage, where it incorporates a diversity-driven memory mechanism that maintains a credible and diverse memory bank, ensuring consistent long-term tracking. Extensive experiments demonstrate that ReSurgSAM2 achieves substantial improvements in accuracy and efficiency compared to existing methods, operating in real-time at 61.2 FPS. Our code and datasets will be available at https://github.com/jinlab-imvr/ReSurgSAM2.
ToolTipNet: A Segmentation-Driven Deep Learning Baseline for Surgical Instrument Tip Detection
Apr 13, 2025Abstract:In robot-assisted laparoscopic radical prostatectomy (RALP), the location of the instrument tip is important to register the ultrasound frame with the laparoscopic camera frame. A long-standing limitation is that the instrument tip position obtained from the da Vinci API is inaccurate and requires hand-eye calibration. Thus, directly computing the position of the tool tip in the camera frame using the vision-based method becomes an attractive solution. Besides, surgical instrument tip detection is the key component of other tasks, like surgical skill assessment and surgery automation. However, this task is challenging due to the small size of the tool tip and the articulation of the surgical instrument. Surgical instrument segmentation becomes relatively easy due to the emergence of the Segmentation Foundation Model, i.e., Segment Anything. Based on this advancement, we explore the deep learning-based surgical instrument tip detection approach that takes the part-level instrument segmentation mask as input. Comparison experiments with a hand-crafted image-processing approach demonstrate the superiority of the proposed method on simulated and real datasets.
Dynamic Allocation Hypernetwork with Adaptive Model Recalibration for Federated Continual Learning
Mar 25, 2025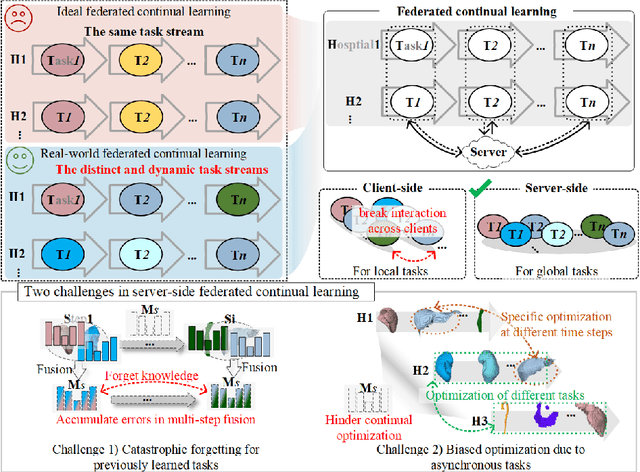

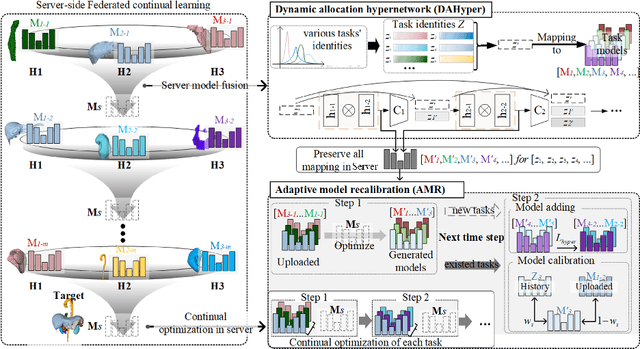
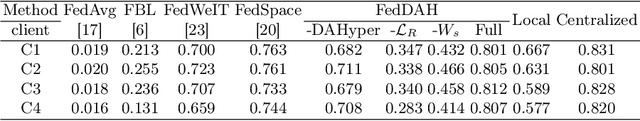
Abstract:Federated continual learning (FCL) offers an emerging pattern to facilitate the applicability of federated learning (FL) in real-world scenarios, where tasks evolve dynamically and asynchronously across clients, especially in medical scenario. Existing server-side FCL methods in nature domain construct a continually learnable server model by client aggregation on all-involved tasks. However, they are challenged by: (1) Catastrophic forgetting for previously learned tasks, leading to error accumulation in server model, making it difficult to sustain comprehensive knowledge across all tasks. (2) Biased optimization due to asynchronous tasks handled across different clients, leading to the collision of optimization targets of different clients at the same time steps. In this work, we take the first step to propose a novel server-side FCL pattern in medical domain, Dynamic Allocation Hypernetwork with adaptive model recalibration (FedDAH). It is to facilitate collaborative learning under the distinct and dynamic task streams across clients. To alleviate the catastrophic forgetting, we propose a dynamic allocation hypernetwork (DAHyper) where a continually updated hypernetwork is designed to manage the mapping between task identities and their associated model parameters, enabling the dynamic allocation of the model across clients. For the biased optimization, we introduce a novel adaptive model recalibration (AMR) to incorporate the candidate changes of historical models into current server updates, and assign weights to identical tasks across different time steps based on the similarity for continual optimization. Extensive experiments on the AMOS dataset demonstrate the superiority of our FedDAH to other FCL methods on sites with different task streams. The code is available:https://github.com/jinlab-imvr/FedDAH.
SurgRAW: Multi-Agent Workflow with Chain-of-Thought Reasoning for Surgical Intelligence
Mar 13, 2025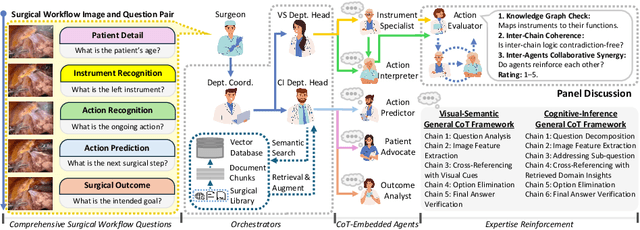
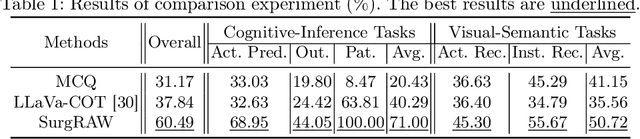
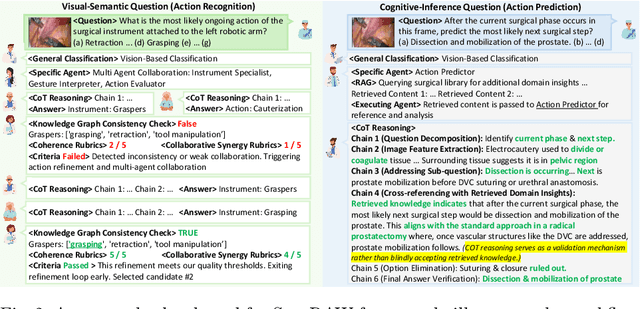
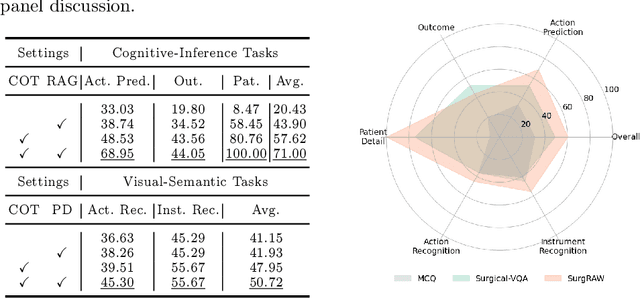
Abstract:Integration of Vision-Language Models (VLMs) in surgical intelligence is hindered by hallucinations, domain knowledge gaps, and limited understanding of task interdependencies within surgical scenes, undermining clinical reliability. While recent VLMs demonstrate strong general reasoning and thinking capabilities, they still lack the domain expertise and task-awareness required for precise surgical scene interpretation. Although Chain-of-Thought (CoT) can structure reasoning more effectively, current approaches rely on self-generated CoT steps, which often exacerbate inherent domain gaps and hallucinations. To overcome this, we present SurgRAW, a CoT-driven multi-agent framework that delivers transparent, interpretable insights for most tasks in robotic-assisted surgery. By employing specialized CoT prompts across five tasks: instrument recognition, action recognition, action prediction, patient data extraction, and outcome assessment, SurgRAW mitigates hallucinations through structured, domain-aware reasoning. Retrieval-Augmented Generation (RAG) is also integrated to external medical knowledge to bridge domain gaps and improve response reliability. Most importantly, a hierarchical agentic system ensures that CoT-embedded VLM agents collaborate effectively while understanding task interdependencies, with a panel discussion mechanism promotes logical consistency. To evaluate our method, we introduce SurgCoTBench, the first reasoning-based dataset with structured frame-level annotations. With comprehensive experiments, we demonstrate the effectiveness of proposed SurgRAW with 29.32% accuracy improvement over baseline VLMs on 12 robotic procedures, achieving the state-of-the-art performance and advancing explainable, trustworthy, and autonomous surgical assistance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge