Shangqing Lyu
PathRWKV: Enabling Whole Slide Prediction with Recurrent-Transformer
Mar 05, 2025


Abstract:Pathological diagnosis plays a critical role in clinical practice, where the whole slide images (WSIs) are widely applied. Through a two-stage paradigm, recent deep learning approaches enhance the WSI analysis with tile-level feature extracting and slide-level feature modeling. Current Transformer models achieved improvement in the efficiency and accuracy to previous multiple instance learning based approaches. However, three core limitations persist, as they do not: (1) robustly address the modeling on variable scales for different slides, (2) effectively balance model complexity and data availability, and (3) balance training efficiency and inference performance. To explicitly address them, we propose a novel model for slide modeling, PathRWKV. Via a recurrent structure, we enable the model for dynamic perceptible tiles in slide-level modeling, which novelly enables the prediction on all tiles in the inference stage. Moreover, we employ linear attention instead of conventional matrix multiplication attention to reduce model complexity and overfitting problem. Lastly, we hinge multi-task learning to enable modeling on versatile tasks simultaneously, improving training efficiency, and asynchronous structure design to draw an effective conclusion on all tiles during inference, enhancing inference performance. Experimental results suggest that PathRWKV outperforms the current state-of-the-art methods in various downstream tasks on multiple datasets. The code and datasets are publicly available.
UnPuzzle: A Unified Framework for Pathology Image Analysis
Mar 05, 2025Abstract:Pathology image analysis plays a pivotal role in medical diagnosis, with deep learning techniques significantly advancing diagnostic accuracy and research. While numerous studies have been conducted to address specific pathological tasks, the lack of standardization in pre-processing methods and model/database architectures complicates fair comparisons across different approaches. This highlights the need for a unified pipeline and comprehensive benchmarks to enable consistent evaluation and accelerate research progress. In this paper, we present UnPuzzle, a novel and unified framework for pathological AI research that covers a broad range of pathology tasks with benchmark results. From high-level to low-level, upstream to downstream tasks, UnPuzzle offers a modular pipeline that encompasses data pre-processing, model composition,taskconfiguration,andexperimentconduction.Specifically, it facilitates efficient benchmarking for both Whole Slide Images (WSIs) and Region of Interest (ROI) tasks. Moreover, the framework supports variouslearningparadigms,includingself-supervisedlearning,multi-task learning,andmulti-modallearning,enablingcomprehensivedevelopment of pathology AI models. Through extensive benchmarking across multiple datasets, we demonstrate the effectiveness of UnPuzzle in streamlining pathology AI research and promoting reproducibility. We envision UnPuzzle as a cornerstone for future advancements in pathology AI, providing a more accessible, transparent, and standardized approach to model evaluation. The UnPuzzle repository is publicly available at https://github.com/Puzzle-AI/UnPuzzle.
PuzzleTuning: Explicitly Bridge Pathological and Natural Image with Puzzles
Nov 12, 2023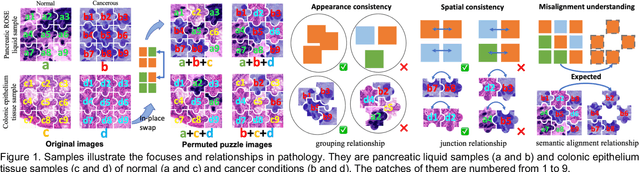
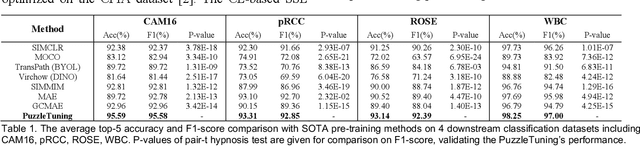
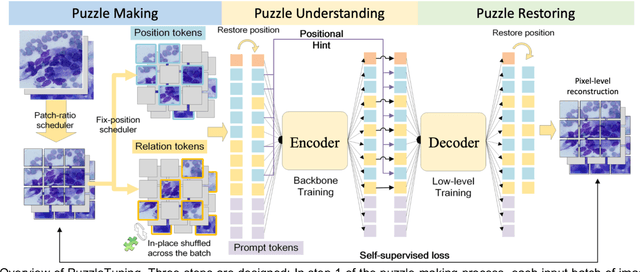
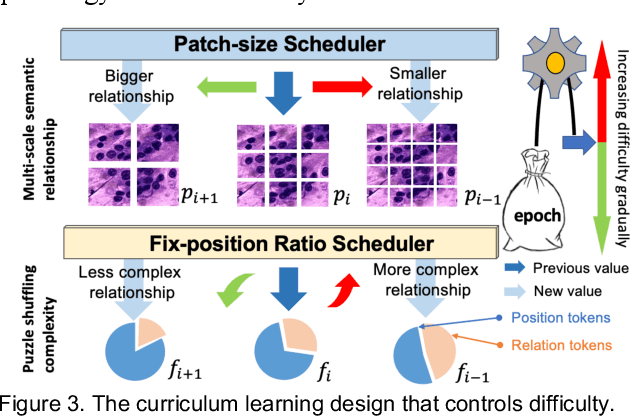
Abstract:Pathological image analysis is a crucial field in computer vision. Due to the annotation scarcity in the pathological field, recently, most of the works leverage self-supervised learning (SSL) trained on unlabeled pathological images, hoping to mine the main representation automatically. However, there are two core defects in SSL-based pathological pre-training: (1) they do not explicitly explore the essential focuses of the pathological field, and (2) they do not effectively bridge with and thus take advantage of the large natural image domain. To explicitly address them, we propose our large-scale PuzzleTuning framework, containing the following innovations. Firstly, we identify three task focuses that can effectively bridge pathological and natural domains: appearance consistency, spatial consistency, and misalignment understanding. Secondly, we devise a multiple puzzle restoring task to explicitly pre-train the model with these focuses. Thirdly, for the existing large domain gap between natural and pathological fields, we introduce an explicit prompt-tuning process to incrementally integrate the domain-specific knowledge with the natural knowledge. Additionally, we design a curriculum-learning training strategy that regulates the task difficulty, making the model fit the complex multiple puzzle restoring task adaptively. Experimental results show that our PuzzleTuning framework outperforms the previous SOTA methods in various downstream tasks on multiple datasets. The code, demo, and pre-trained weights are available at https://github.com/sagizty/PuzzleTuning.
CPIA Dataset: A Comprehensive Pathological Image Analysis Dataset for Self-supervised Learning Pre-training
Oct 27, 2023Abstract:Pathological image analysis is a crucial field in computer-aided diagnosis, where deep learning is widely applied. Transfer learning using pre-trained models initialized on natural images has effectively improved the downstream pathological performance. However, the lack of sophisticated domain-specific pathological initialization hinders their potential. Self-supervised learning (SSL) enables pre-training without sample-level labels, which has great potential to overcome the challenge of expensive annotations. Thus, studies focusing on pathological SSL pre-training call for a comprehensive and standardized dataset, similar to the ImageNet in computer vision. This paper presents the comprehensive pathological image analysis (CPIA) dataset, a large-scale SSL pre-training dataset combining 103 open-source datasets with extensive standardization. The CPIA dataset contains 21,427,877 standardized images, covering over 48 organs/tissues and about 100 kinds of diseases, which includes two main data types: whole slide images (WSIs) and characteristic regions of interest (ROIs). A four-scale WSI standardization process is proposed based on the uniform resolution in microns per pixel (MPP), while the ROIs are divided into three scales artificially. This multi-scale dataset is built with the diagnosis habits under the supervision of experienced senior pathologists. The CPIA dataset facilitates a comprehensive pathological understanding and enables pattern discovery explorations. Additionally, to launch the CPIA dataset, several state-of-the-art (SOTA) baselines of SSL pre-training and downstream evaluation are specially conducted. The CPIA dataset along with baselines is available at https://github.com/zhanglab2021/CPIA_Dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge