Guanglei Zhang
Diff-FMT: Diffusion Models for Fluorescence Molecular Tomography
Oct 09, 2024Abstract:Fluorescence molecular tomography (FMT) is a real-time, noninvasive optical imaging technology that plays a significant role in biomedical research. Nevertheless, the ill-posedness of the inverse problem poses huge challenges in FMT reconstructions. Previous various deep learning algorithms have been extensively explored to address the critical issues, but they remain faces the challenge of high data dependency with poor image quality. In this paper, we, for the first time, propose a FMT reconstruction method based on a denoising diffusion probabilistic model (DDPM), termed Diff-FMT, which is capable of obtaining high-quality reconstructed images from noisy images. Specifically, we utilize the noise addition mechanism of DDPM to generate diverse training samples. Through the step-by-step probability sampling mechanism in the inverse process, we achieve fine-grained reconstruction of the image, avoiding issues such as loss of image detail that can occur with end-to-end deep-learning methods. Additionally, we introduce the fluorescence signals as conditional information in the model training to sample a reconstructed image that is highly consistent with the input fluorescence signals from the noisy images. Numerous experimental results show that Diff-FMT can achieve high-resolution reconstruction images without relying on large-scale datasets compared with other cutting-edge algorithms.
Overcoming Pathology Image Data Deficiency: Generating Images from Pathological Transformation Process
Nov 21, 2023Abstract:Histopathology serves as the gold standard for medical diagnosis but faces application limitations due to the shortage of medical resources. Leveraging deep learning, computer-aided diagnosis has the potential to alleviate the pathologist scarcity and provide timely clinical analysis. However, developing a reliable model generally necessitates substantial data for training, which is challenging in pathological field. In response, we propose an adaptive depth-controlled bidirectional diffusion (ADBD) network for image data generation. The domain migration approach can work with small trainset and overcome the diffusion overfitting by source information guidance. Specifically, we developed a hybrid attention strategy to blend global and local attention priorities, which guides the bidirectional diffusion and ensures the migration success. In addition, we developed the adaptive depth-controlled strategy to simulate physiological transformations, capable of yielding unlimited cross-domain intermediate images with corresponding soft labels. ADBD is effective for overcoming pathological image data deficiency and supportable for further pathology-related research.
PuzzleTuning: Explicitly Bridge Pathological and Natural Image with Puzzles
Nov 12, 2023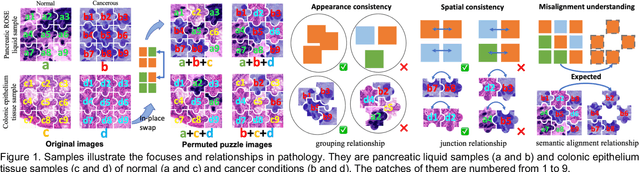
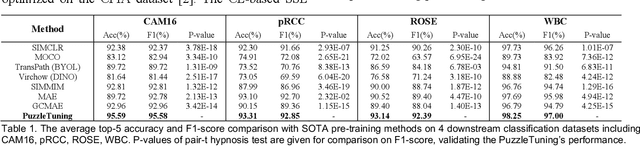
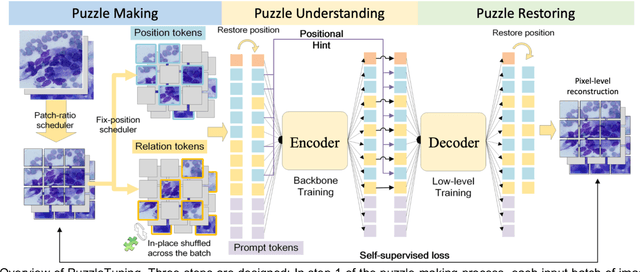
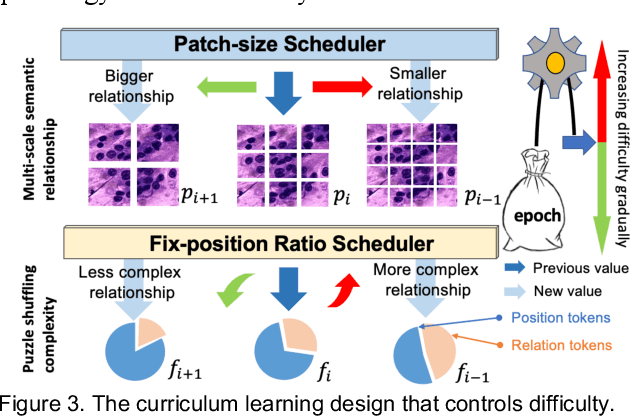
Abstract:Pathological image analysis is a crucial field in computer vision. Due to the annotation scarcity in the pathological field, recently, most of the works leverage self-supervised learning (SSL) trained on unlabeled pathological images, hoping to mine the main representation automatically. However, there are two core defects in SSL-based pathological pre-training: (1) they do not explicitly explore the essential focuses of the pathological field, and (2) they do not effectively bridge with and thus take advantage of the large natural image domain. To explicitly address them, we propose our large-scale PuzzleTuning framework, containing the following innovations. Firstly, we identify three task focuses that can effectively bridge pathological and natural domains: appearance consistency, spatial consistency, and misalignment understanding. Secondly, we devise a multiple puzzle restoring task to explicitly pre-train the model with these focuses. Thirdly, for the existing large domain gap between natural and pathological fields, we introduce an explicit prompt-tuning process to incrementally integrate the domain-specific knowledge with the natural knowledge. Additionally, we design a curriculum-learning training strategy that regulates the task difficulty, making the model fit the complex multiple puzzle restoring task adaptively. Experimental results show that our PuzzleTuning framework outperforms the previous SOTA methods in various downstream tasks on multiple datasets. The code, demo, and pre-trained weights are available at https://github.com/sagizty/PuzzleTuning.
CPIA Dataset: A Comprehensive Pathological Image Analysis Dataset for Self-supervised Learning Pre-training
Oct 27, 2023Abstract:Pathological image analysis is a crucial field in computer-aided diagnosis, where deep learning is widely applied. Transfer learning using pre-trained models initialized on natural images has effectively improved the downstream pathological performance. However, the lack of sophisticated domain-specific pathological initialization hinders their potential. Self-supervised learning (SSL) enables pre-training without sample-level labels, which has great potential to overcome the challenge of expensive annotations. Thus, studies focusing on pathological SSL pre-training call for a comprehensive and standardized dataset, similar to the ImageNet in computer vision. This paper presents the comprehensive pathological image analysis (CPIA) dataset, a large-scale SSL pre-training dataset combining 103 open-source datasets with extensive standardization. The CPIA dataset contains 21,427,877 standardized images, covering over 48 organs/tissues and about 100 kinds of diseases, which includes two main data types: whole slide images (WSIs) and characteristic regions of interest (ROIs). A four-scale WSI standardization process is proposed based on the uniform resolution in microns per pixel (MPP), while the ROIs are divided into three scales artificially. This multi-scale dataset is built with the diagnosis habits under the supervision of experienced senior pathologists. The CPIA dataset facilitates a comprehensive pathological understanding and enables pattern discovery explorations. Additionally, to launch the CPIA dataset, several state-of-the-art (SOTA) baselines of SSL pre-training and downstream evaluation are specially conducted. The CPIA dataset along with baselines is available at https://github.com/zhanglab2021/CPIA_Dataset.
CellMix: A General Instance Relationship based Method for Data Augmentation Towards Pathology Image Analysis
Jan 27, 2023
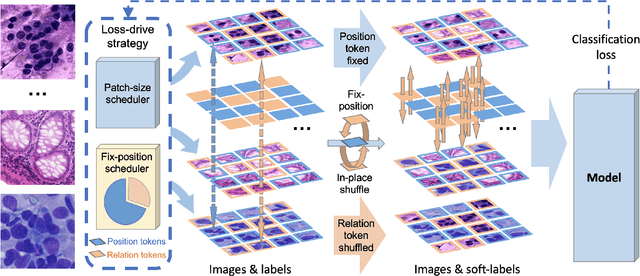
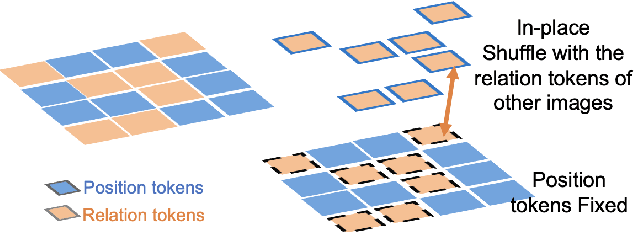
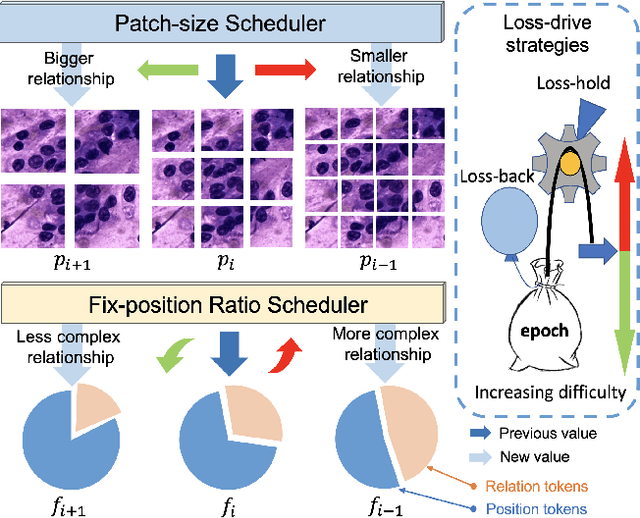
Abstract:Pathology image analysis crucially relies on the availability and quality of annotated pathological samples, which are very difficult to collect and need lots of human effort. To address this issue, beyond traditional preprocess data augmentation methods, mixing-based approaches are effective and practical. However, previous mixing-based data augmentation methods do not thoroughly explore the essential characteristics of pathology images, including the local specificity, global distribution, and inner/outer-sample instance relationship. To further understand the pathology characteristics and make up effective pseudo samples, we propose the CellMix framework with a novel distribution-based in-place shuffle strategy. We split the images into patches with respect to the granularity of pathology instances and do the shuffle process across the same batch. In this way, we generate new samples while keeping the absolute relationship of pathology instances intact. Furthermore, to deal with the perturbations and distribution-based noise, we devise a loss-drive strategy inspired by curriculum learning during the training process, making the model fit the augmented data adaptively. It is worth mentioning that we are the first to explore data augmentation techniques in the pathology image field. Experiments show SOTA results on 7 different datasets. We conclude that this novel instance relationship-based strategy can shed light on general data augmentation for pathology image analysis. The code is available at https://github.com/sagizty/CellMix.
Shuffle Instances-based Vision Transformer for Pancreatic Cancer ROSE Image Classification
Aug 14, 2022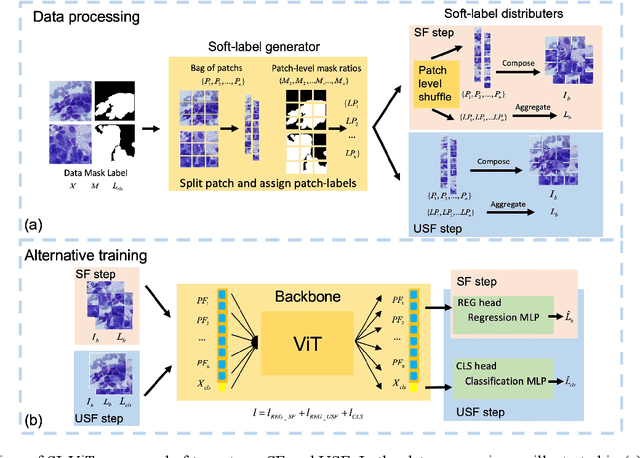
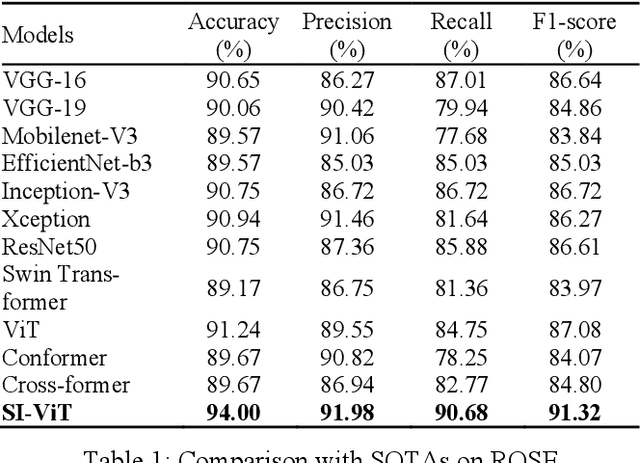
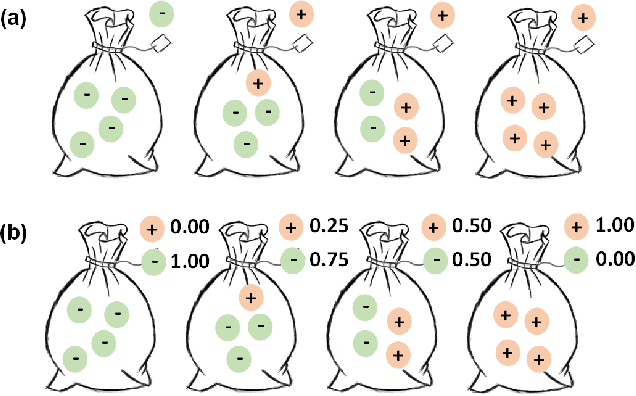
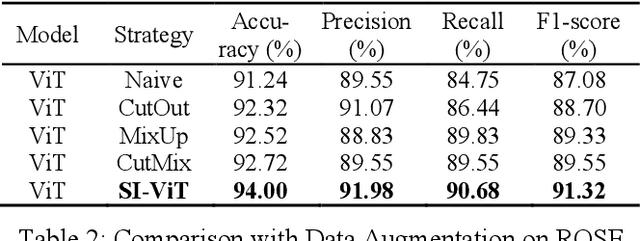
Abstract:The rapid on-site evaluation (ROSE) technique can signifi-cantly accelerate the diagnosis of pancreatic cancer by im-mediately analyzing the fast-stained cytopathological images. Computer-aided diagnosis (CAD) can potentially address the shortage of pathologists in ROSE. However, the cancerous patterns vary significantly between different samples, making the CAD task extremely challenging. Besides, the ROSE images have complicated perturbations regarding color distribution, brightness, and contrast due to different staining qualities and various acquisition device types. To address these challenges, we proposed a shuffle instances-based Vision Transformer (SI-ViT) approach, which can reduce the perturbations and enhance the modeling among the instances. With the regrouped bags of shuffle instances and their bag-level soft labels, the approach utilizes a regression head to make the model focus on the cells rather than various perturbations. Simultaneously, combined with a classification head, the model can effectively identify the general distributive patterns among different instances. The results demonstrate significant improvements in the classification accuracy with more accurate attention regions, indicating that the diverse patterns of ROSE images are effectively extracted, and the complicated perturbations are significantly reduced. It also suggests that the SI-ViT has excellent potential in analyzing cytopathological images. The code and experimental results are available at https://github.com/sagizty/MIL-SI.
Pancreatic Cancer ROSE Image Classification Based on Multiple Instance Learning with Shuffle Instances
Jun 07, 2022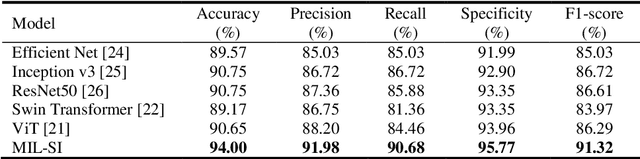
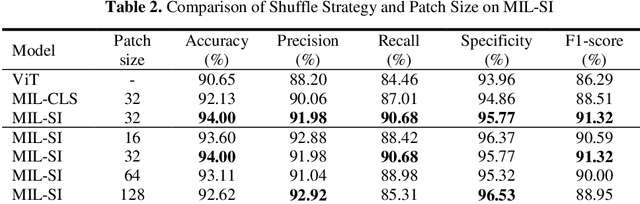
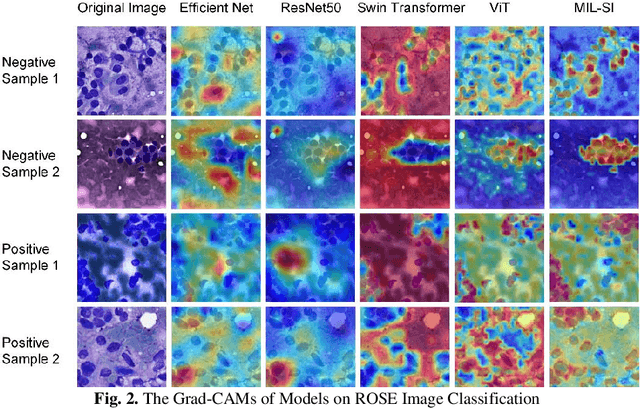
Abstract:The rapid on-site evaluation (ROSE) technique can significantly ac-celerate the diagnostic workflow of pancreatic cancer by immediately analyzing the fast-stained cytopathological images with on-site pathologists. Computer-aided diagnosis (CAD) using the deep learning method has the potential to solve the problem of insufficient pathology staffing. However, the cancerous patterns of ROSE images vary greatly between different samples, making the CAD task extremely challenging. Besides, due to different staining qualities and various types of acquisition devices, the ROSE images also have compli-cated perturbations in terms of color distribution, brightness, and contrast. To address these challenges, we proposed a novel multiple instance learning (MIL) approach using shuffle patches containing the instances, which adopts the patch-based learning strategy of Vision Transformers. With the re-grouped bags of shuffle instances and their bag-level soft labels, the approach utilizes a MIL head to make the model focus on the features from the pancreatic cancer cells, rather than that from various perturbations in ROSE images. Simultaneously, combined with a classification head, the model can effectively identify the gen-eral distributive patterns across different instances. The results demonstrate the significant improvements in the classification accuracy with more accurate at-tention regions, indicating that the diverse patterns of ROSE images are effec-tively extracted, and the complicated perturbations of ROSE images are signifi-cantly eliminated. It also suggests that the MIL with shuffle instances has great potential in the analysis of cytopathological images.
MSHT: Multi-stage Hybrid Transformer for the ROSE Image Analysis of Pancreatic Cancer
Dec 27, 2021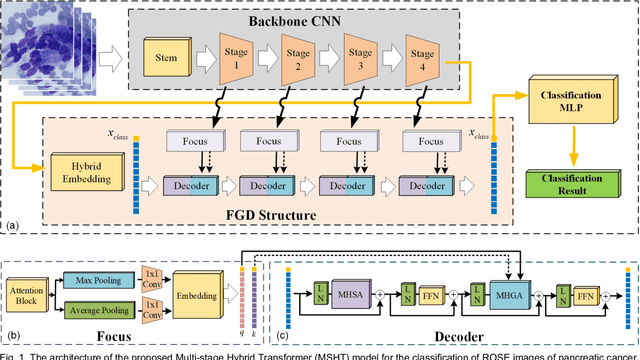
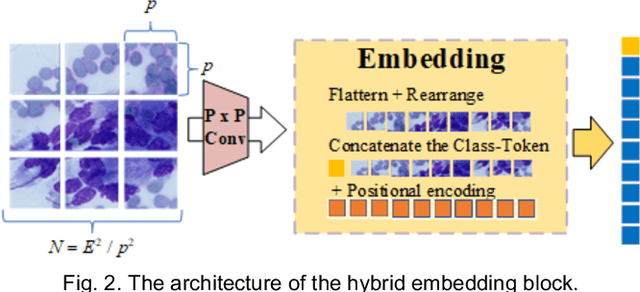
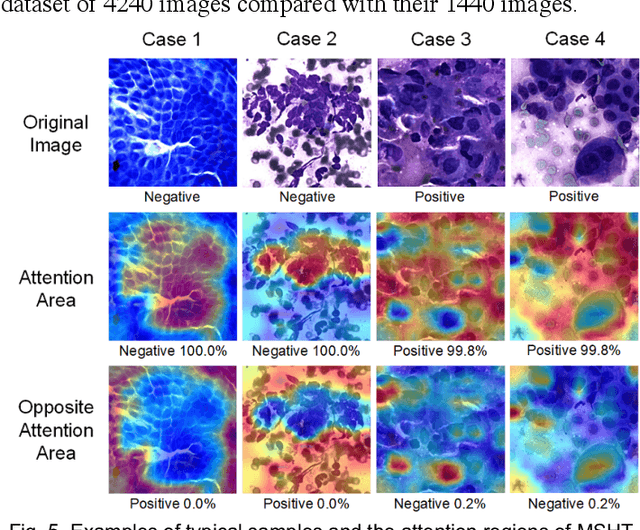

Abstract:Pancreatic cancer is one of the most malignant cancers in the world, which deteriorates rapidly with very high mortality. The rapid on-site evaluation (ROSE) technique innovates the workflow by immediately analyzing the fast stained cytopathological images with on-site pathologists, which enables faster diagnosis in this time-pressured process. However, the wider expansion of ROSE diagnosis has been hindered by the lack of experienced pathologists. To overcome this problem, we propose a hybrid high-performance deep learning model to enable the automated workflow, thus freeing the occupation of the valuable time of pathologists. By firstly introducing the Transformer block into this field with our particular multi-stage hybrid design, the spatial features generated by the convolutional neural network (CNN) significantly enhance the Transformer global modeling. Turning multi-stage spatial features as global attention guidance, this design combines the robustness from the inductive bias of CNN with the sophisticated global modeling power of Transformer. A dataset of 4240 ROSE images is collected to evaluate the method in this unexplored field. The proposed multi-stage hybrid Transformer (MSHT) achieves 95.68% in classification accuracy, which is distinctively higher than the state-of-the-art models. Facing the need for interpretability, MSHT outperforms its counterparts with more accurate attention regions. The results demonstrate that the MSHT can distinguish cancer samples accurately at an unprecedented image scale, laying the foundation for deploying automatic decision systems and enabling the expansion of ROSE in clinical practice. The code and records are available at: https://github.com/sagizty/Multi-Stage-Hybrid-Transformer.
Prior Attention Network for Multi-Lesion Segmentation in Medical Images
Oct 10, 2021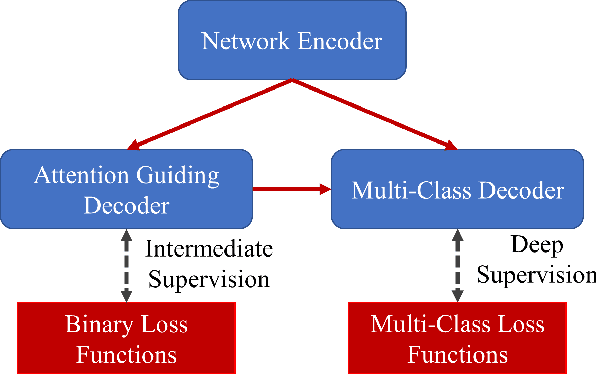
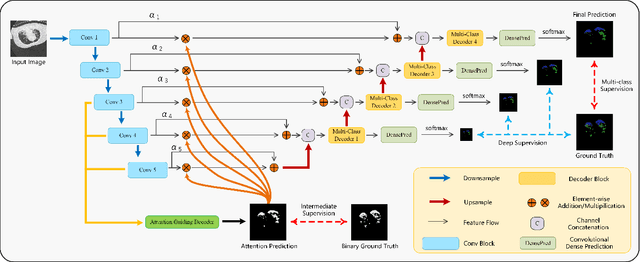
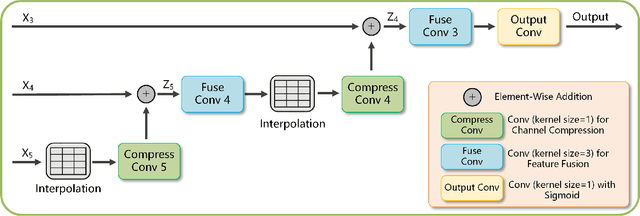
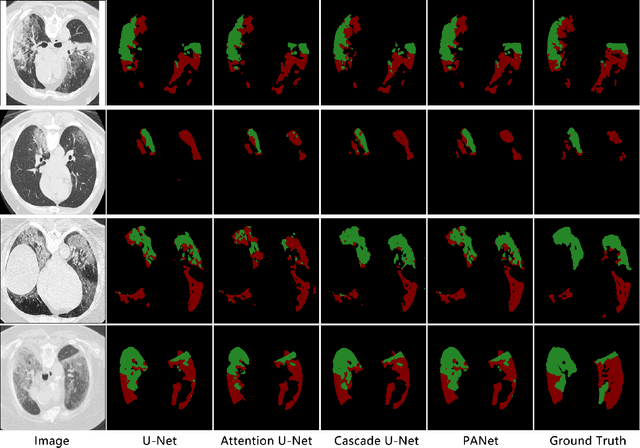
Abstract:The accurate segmentation of multiple types of lesions from adjacent tissues in medical images is significant in clinical practice. Convolutional neural networks (CNNs) based on the coarse-to-fine strategy have been widely used in this field. However, multi-lesion segmentation remains to be challenging due to the uncertainty in size, contrast, and high interclass similarity of tissues. In addition, the commonly adopted cascaded strategy is rather demanding in terms of hardware, which limits the potential of clinical deployment. To address the problems above,we propose a novel Prior Attention Network (PANet) that follows the coarse-to-fine strategy to perform multi-lesion segmentation in medical images. The proposed network achieves the two steps of segmentation in a single network by inserting lesion-related spatial attention mechanism in the network. Further, we also propose the intermediate supervision strategy for generating lesion-related attention to acquire the regions of interest (ROIs), which accelerates the convergence and obviously improves the segmentation performance. We have investigated the proposed segmentation framework in two applications: 2D segmentation of multiple lung infections in lung CT slices and 3D segmentation of multiple lesions in brain MRIs. Experimental results show that in both 2D and 3D segmentation tasks our proposed network achieves better performance with less computational cost compared with cascaded networks. The proposed network can be regarded as a universal solution to multi-lesion segmentation in both 2D and 3D tasks. The source code is available at: https://github.com/hsiangyuzhao/PANet.
D2A U-Net: Automatic Segmentation of COVID-19 Lesions from CT Slices with Dilated Convolution and Dual Attention Mechanism
Feb 10, 2021



Abstract:Coronavirus Disease 2019 (COVID-19) has caused great casualties and becomes almost the most urgent public health events worldwide. Computed tomography (CT) is a significant screening tool for COVID-19 infection, and automated segmentation of lung infection in COVID-19 CT images will greatly assist diagnosis and health care of patients. However, accurate and automatic segmentation of COVID-19 lung infections remains to be challenging. In this paper we propose a dilated dual attention U-Net (D2A U-Net) for COVID-19 lesion segmentation in CT slices based on dilated convolution and a novel dual attention mechanism to address the issues above. We introduce a dilated convolution module in model decoder to achieve large receptive field, which refines decoding process and contributes to segmentation accuracy. Also, we present a dual attention mechanism composed of two attention modules which are inserted to skip connection and model decoder respectively. The dual attention mechanism is utilized to refine feature maps and reduce semantic gap between different levels of the model. The proposed method has been evaluated on open-source dataset and outperforms cutting edges methods in semantic segmentation. Our proposed D2A U-Net with pretrained encoder achieves a Dice score of 0.7298 and recall score of 0.7071. Besides, we also build a simplified D2A U-Net without pretrained encoder to provide a fair comparison with other models trained from scratch, which still outperforms popular U-Net family models with a Dice score of 0.7047 and recall score of 0.6626. Our experiment results have shown that by introducing dilated convolution and dual attention mechanism, the number of false positives is significantly reduced, which improves sensitivity to COVID-19 lesions and subsequently brings significant increase to Dice score.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge