CellMix: A General Instance Relationship based Method for Data Augmentation Towards Pathology Image Analysis
Paper and Code
Jan 27, 2023
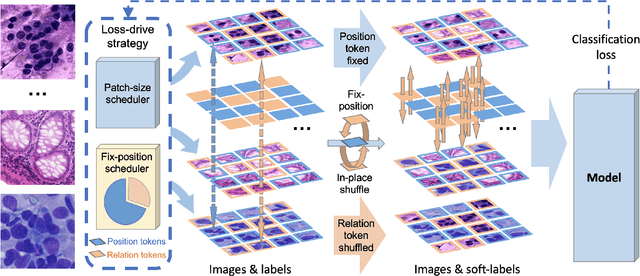
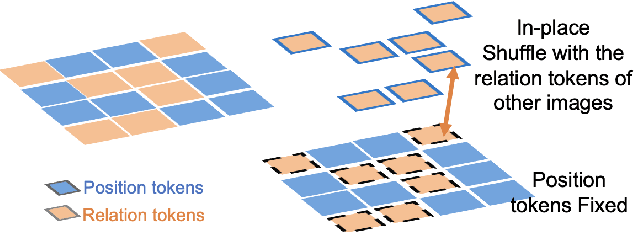
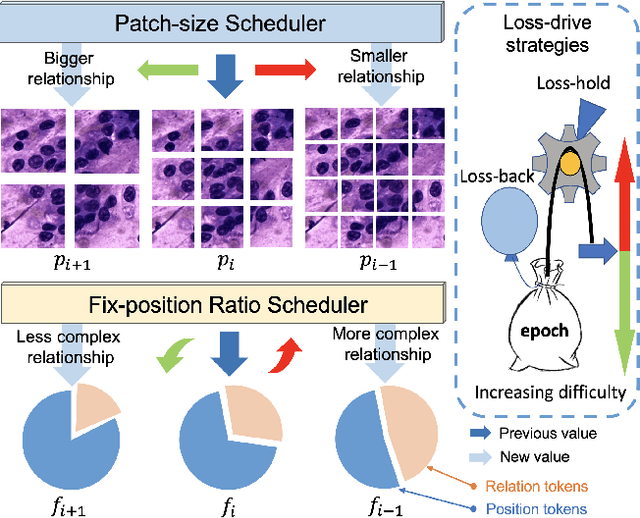
Pathology image analysis crucially relies on the availability and quality of annotated pathological samples, which are very difficult to collect and need lots of human effort. To address this issue, beyond traditional preprocess data augmentation methods, mixing-based approaches are effective and practical. However, previous mixing-based data augmentation methods do not thoroughly explore the essential characteristics of pathology images, including the local specificity, global distribution, and inner/outer-sample instance relationship. To further understand the pathology characteristics and make up effective pseudo samples, we propose the CellMix framework with a novel distribution-based in-place shuffle strategy. We split the images into patches with respect to the granularity of pathology instances and do the shuffle process across the same batch. In this way, we generate new samples while keeping the absolute relationship of pathology instances intact. Furthermore, to deal with the perturbations and distribution-based noise, we devise a loss-drive strategy inspired by curriculum learning during the training process, making the model fit the augmented data adaptively. It is worth mentioning that we are the first to explore data augmentation techniques in the pathology image field. Experiments show SOTA results on 7 different datasets. We conclude that this novel instance relationship-based strategy can shed light on general data augmentation for pathology image analysis. The code is available at https://github.com/sagizty/CellMix.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge