Nan Ying
PuzzleTuning: Explicitly Bridge Pathological and Natural Image with Puzzles
Nov 12, 2023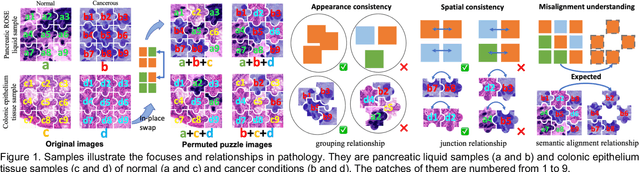
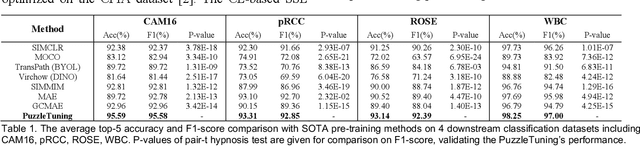
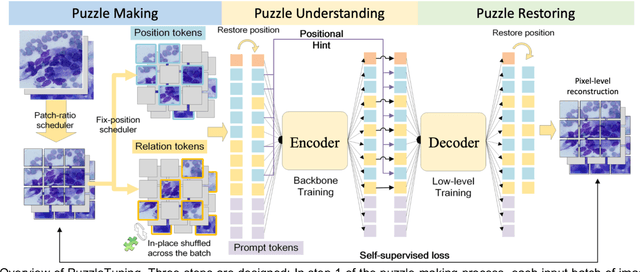
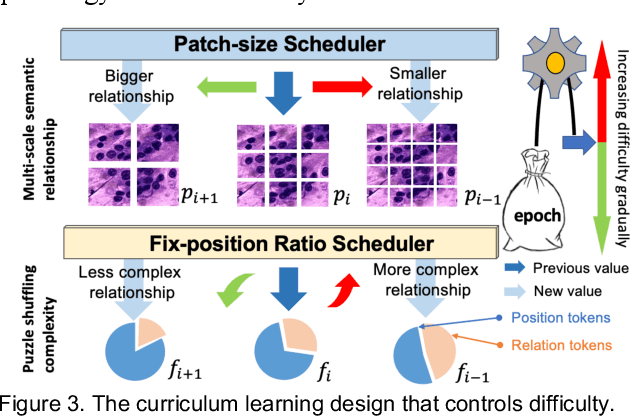
Abstract:Pathological image analysis is a crucial field in computer vision. Due to the annotation scarcity in the pathological field, recently, most of the works leverage self-supervised learning (SSL) trained on unlabeled pathological images, hoping to mine the main representation automatically. However, there are two core defects in SSL-based pathological pre-training: (1) they do not explicitly explore the essential focuses of the pathological field, and (2) they do not effectively bridge with and thus take advantage of the large natural image domain. To explicitly address them, we propose our large-scale PuzzleTuning framework, containing the following innovations. Firstly, we identify three task focuses that can effectively bridge pathological and natural domains: appearance consistency, spatial consistency, and misalignment understanding. Secondly, we devise a multiple puzzle restoring task to explicitly pre-train the model with these focuses. Thirdly, for the existing large domain gap between natural and pathological fields, we introduce an explicit prompt-tuning process to incrementally integrate the domain-specific knowledge with the natural knowledge. Additionally, we design a curriculum-learning training strategy that regulates the task difficulty, making the model fit the complex multiple puzzle restoring task adaptively. Experimental results show that our PuzzleTuning framework outperforms the previous SOTA methods in various downstream tasks on multiple datasets. The code, demo, and pre-trained weights are available at https://github.com/sagizty/PuzzleTuning.
CPIA Dataset: A Comprehensive Pathological Image Analysis Dataset for Self-supervised Learning Pre-training
Oct 27, 2023Abstract:Pathological image analysis is a crucial field in computer-aided diagnosis, where deep learning is widely applied. Transfer learning using pre-trained models initialized on natural images has effectively improved the downstream pathological performance. However, the lack of sophisticated domain-specific pathological initialization hinders their potential. Self-supervised learning (SSL) enables pre-training without sample-level labels, which has great potential to overcome the challenge of expensive annotations. Thus, studies focusing on pathological SSL pre-training call for a comprehensive and standardized dataset, similar to the ImageNet in computer vision. This paper presents the comprehensive pathological image analysis (CPIA) dataset, a large-scale SSL pre-training dataset combining 103 open-source datasets with extensive standardization. The CPIA dataset contains 21,427,877 standardized images, covering over 48 organs/tissues and about 100 kinds of diseases, which includes two main data types: whole slide images (WSIs) and characteristic regions of interest (ROIs). A four-scale WSI standardization process is proposed based on the uniform resolution in microns per pixel (MPP), while the ROIs are divided into three scales artificially. This multi-scale dataset is built with the diagnosis habits under the supervision of experienced senior pathologists. The CPIA dataset facilitates a comprehensive pathological understanding and enables pattern discovery explorations. Additionally, to launch the CPIA dataset, several state-of-the-art (SOTA) baselines of SSL pre-training and downstream evaluation are specially conducted. The CPIA dataset along with baselines is available at https://github.com/zhanglab2021/CPIA_Dataset.
CellMix: A General Instance Relationship based Method for Data Augmentation Towards Pathology Image Analysis
Jan 27, 2023
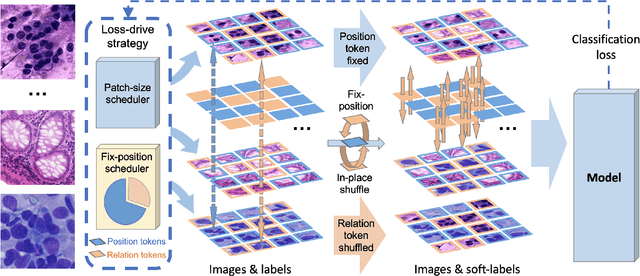
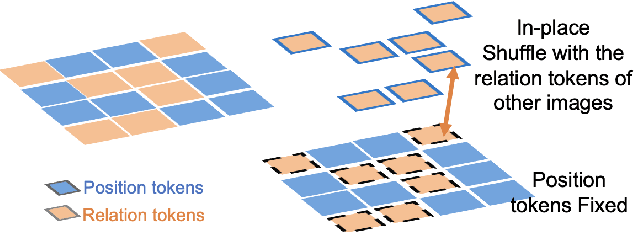
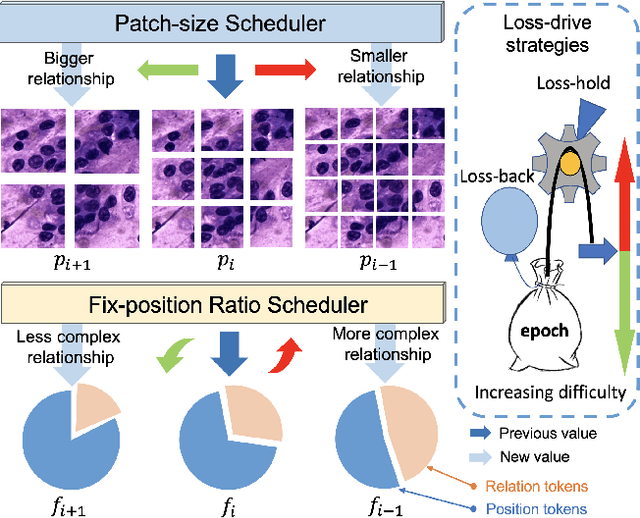
Abstract:Pathology image analysis crucially relies on the availability and quality of annotated pathological samples, which are very difficult to collect and need lots of human effort. To address this issue, beyond traditional preprocess data augmentation methods, mixing-based approaches are effective and practical. However, previous mixing-based data augmentation methods do not thoroughly explore the essential characteristics of pathology images, including the local specificity, global distribution, and inner/outer-sample instance relationship. To further understand the pathology characteristics and make up effective pseudo samples, we propose the CellMix framework with a novel distribution-based in-place shuffle strategy. We split the images into patches with respect to the granularity of pathology instances and do the shuffle process across the same batch. In this way, we generate new samples while keeping the absolute relationship of pathology instances intact. Furthermore, to deal with the perturbations and distribution-based noise, we devise a loss-drive strategy inspired by curriculum learning during the training process, making the model fit the augmented data adaptively. It is worth mentioning that we are the first to explore data augmentation techniques in the pathology image field. Experiments show SOTA results on 7 different datasets. We conclude that this novel instance relationship-based strategy can shed light on general data augmentation for pathology image analysis. The code is available at https://github.com/sagizty/CellMix.
Shuffle Instances-based Vision Transformer for Pancreatic Cancer ROSE Image Classification
Aug 14, 2022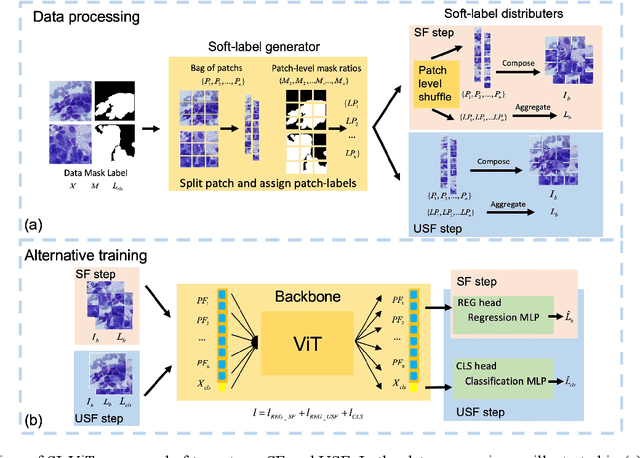
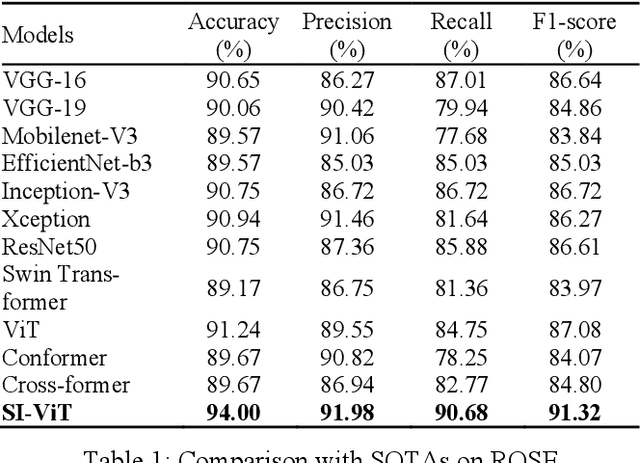
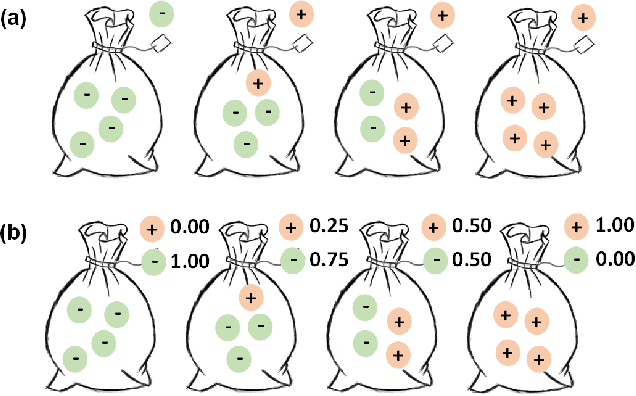
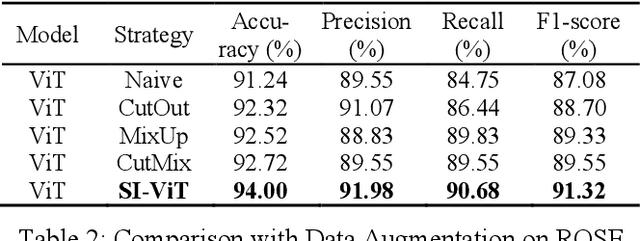
Abstract:The rapid on-site evaluation (ROSE) technique can signifi-cantly accelerate the diagnosis of pancreatic cancer by im-mediately analyzing the fast-stained cytopathological images. Computer-aided diagnosis (CAD) can potentially address the shortage of pathologists in ROSE. However, the cancerous patterns vary significantly between different samples, making the CAD task extremely challenging. Besides, the ROSE images have complicated perturbations regarding color distribution, brightness, and contrast due to different staining qualities and various acquisition device types. To address these challenges, we proposed a shuffle instances-based Vision Transformer (SI-ViT) approach, which can reduce the perturbations and enhance the modeling among the instances. With the regrouped bags of shuffle instances and their bag-level soft labels, the approach utilizes a regression head to make the model focus on the cells rather than various perturbations. Simultaneously, combined with a classification head, the model can effectively identify the general distributive patterns among different instances. The results demonstrate significant improvements in the classification accuracy with more accurate attention regions, indicating that the diverse patterns of ROSE images are effectively extracted, and the complicated perturbations are significantly reduced. It also suggests that the SI-ViT has excellent potential in analyzing cytopathological images. The code and experimental results are available at https://github.com/sagizty/MIL-SI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge