Yufang He
Overcoming Pathology Image Data Deficiency: Generating Images from Pathological Transformation Process
Nov 21, 2023Abstract:Histopathology serves as the gold standard for medical diagnosis but faces application limitations due to the shortage of medical resources. Leveraging deep learning, computer-aided diagnosis has the potential to alleviate the pathologist scarcity and provide timely clinical analysis. However, developing a reliable model generally necessitates substantial data for training, which is challenging in pathological field. In response, we propose an adaptive depth-controlled bidirectional diffusion (ADBD) network for image data generation. The domain migration approach can work with small trainset and overcome the diffusion overfitting by source information guidance. Specifically, we developed a hybrid attention strategy to blend global and local attention priorities, which guides the bidirectional diffusion and ensures the migration success. In addition, we developed the adaptive depth-controlled strategy to simulate physiological transformations, capable of yielding unlimited cross-domain intermediate images with corresponding soft labels. ADBD is effective for overcoming pathological image data deficiency and supportable for further pathology-related research.
PuzzleTuning: Explicitly Bridge Pathological and Natural Image with Puzzles
Nov 12, 2023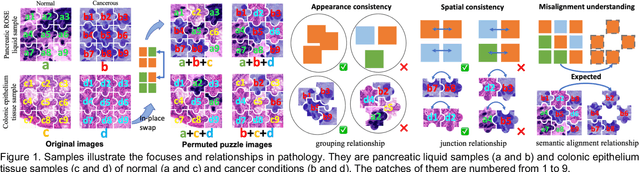
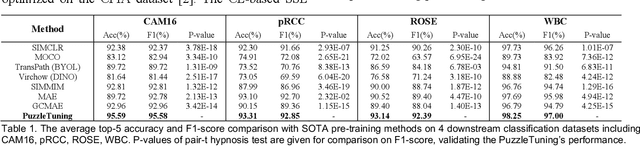
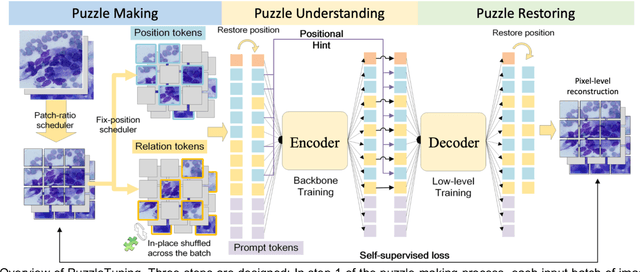
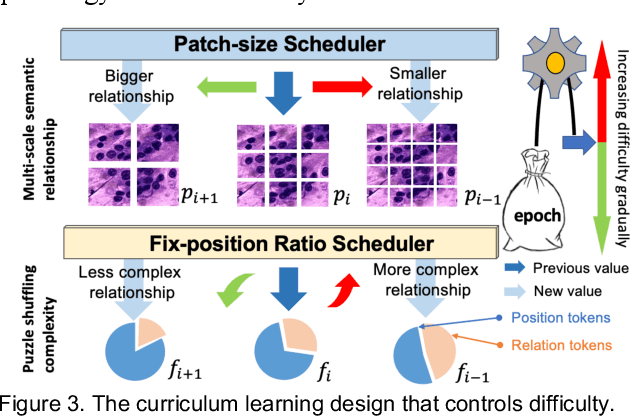
Abstract:Pathological image analysis is a crucial field in computer vision. Due to the annotation scarcity in the pathological field, recently, most of the works leverage self-supervised learning (SSL) trained on unlabeled pathological images, hoping to mine the main representation automatically. However, there are two core defects in SSL-based pathological pre-training: (1) they do not explicitly explore the essential focuses of the pathological field, and (2) they do not effectively bridge with and thus take advantage of the large natural image domain. To explicitly address them, we propose our large-scale PuzzleTuning framework, containing the following innovations. Firstly, we identify three task focuses that can effectively bridge pathological and natural domains: appearance consistency, spatial consistency, and misalignment understanding. Secondly, we devise a multiple puzzle restoring task to explicitly pre-train the model with these focuses. Thirdly, for the existing large domain gap between natural and pathological fields, we introduce an explicit prompt-tuning process to incrementally integrate the domain-specific knowledge with the natural knowledge. Additionally, we design a curriculum-learning training strategy that regulates the task difficulty, making the model fit the complex multiple puzzle restoring task adaptively. Experimental results show that our PuzzleTuning framework outperforms the previous SOTA methods in various downstream tasks on multiple datasets. The code, demo, and pre-trained weights are available at https://github.com/sagizty/PuzzleTuning.
Shuffle Instances-based Vision Transformer for Pancreatic Cancer ROSE Image Classification
Aug 14, 2022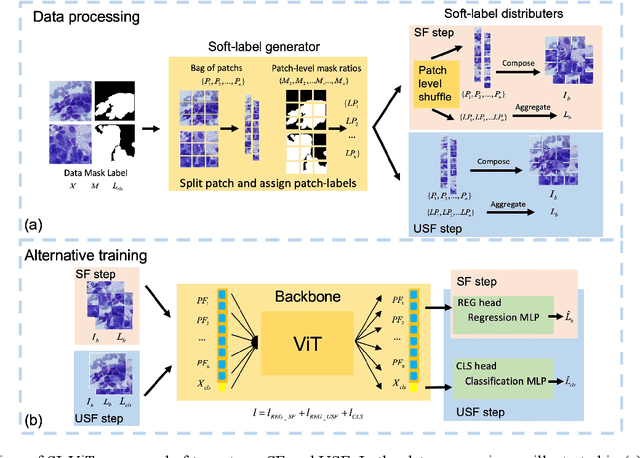
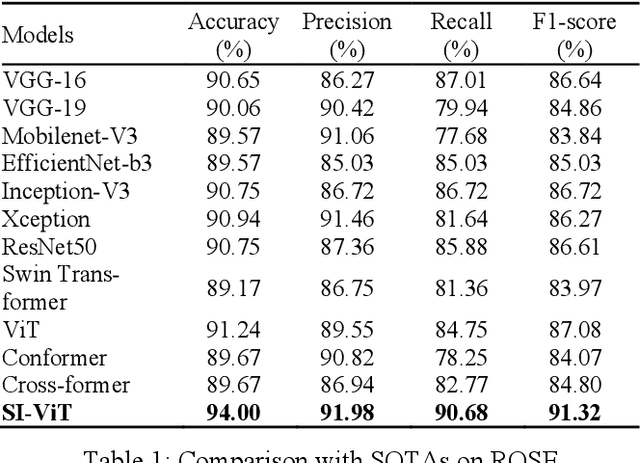
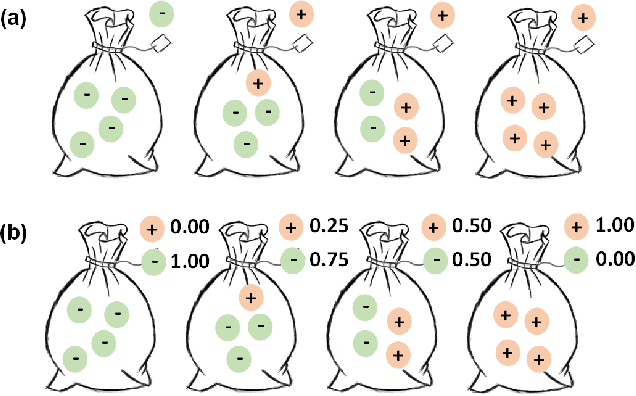
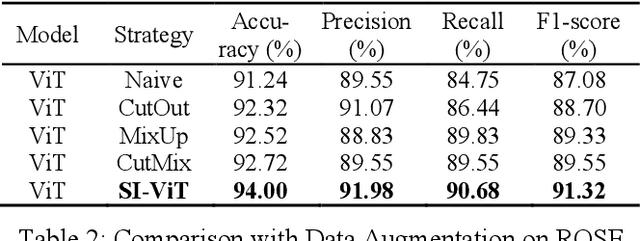
Abstract:The rapid on-site evaluation (ROSE) technique can signifi-cantly accelerate the diagnosis of pancreatic cancer by im-mediately analyzing the fast-stained cytopathological images. Computer-aided diagnosis (CAD) can potentially address the shortage of pathologists in ROSE. However, the cancerous patterns vary significantly between different samples, making the CAD task extremely challenging. Besides, the ROSE images have complicated perturbations regarding color distribution, brightness, and contrast due to different staining qualities and various acquisition device types. To address these challenges, we proposed a shuffle instances-based Vision Transformer (SI-ViT) approach, which can reduce the perturbations and enhance the modeling among the instances. With the regrouped bags of shuffle instances and their bag-level soft labels, the approach utilizes a regression head to make the model focus on the cells rather than various perturbations. Simultaneously, combined with a classification head, the model can effectively identify the general distributive patterns among different instances. The results demonstrate significant improvements in the classification accuracy with more accurate attention regions, indicating that the diverse patterns of ROSE images are effectively extracted, and the complicated perturbations are significantly reduced. It also suggests that the SI-ViT has excellent potential in analyzing cytopathological images. The code and experimental results are available at https://github.com/sagizty/MIL-SI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge