Wenjian Wang
Diff-FMT: Diffusion Models for Fluorescence Molecular Tomography
Oct 09, 2024Abstract:Fluorescence molecular tomography (FMT) is a real-time, noninvasive optical imaging technology that plays a significant role in biomedical research. Nevertheless, the ill-posedness of the inverse problem poses huge challenges in FMT reconstructions. Previous various deep learning algorithms have been extensively explored to address the critical issues, but they remain faces the challenge of high data dependency with poor image quality. In this paper, we, for the first time, propose a FMT reconstruction method based on a denoising diffusion probabilistic model (DDPM), termed Diff-FMT, which is capable of obtaining high-quality reconstructed images from noisy images. Specifically, we utilize the noise addition mechanism of DDPM to generate diverse training samples. Through the step-by-step probability sampling mechanism in the inverse process, we achieve fine-grained reconstruction of the image, avoiding issues such as loss of image detail that can occur with end-to-end deep-learning methods. Additionally, we introduce the fluorescence signals as conditional information in the model training to sample a reconstructed image that is highly consistent with the input fluorescence signals from the noisy images. Numerous experimental results show that Diff-FMT can achieve high-resolution reconstruction images without relying on large-scale datasets compared with other cutting-edge algorithms.
PipeOptim: Ensuring Effective 1F1B Schedule with Optimizer-Dependent Weight Prediction
Dec 05, 2023Abstract:Asynchronous pipeline model parallelism with a "1F1B" (one forward, one backward) schedule generates little bubble overhead and always provides quite a high throughput. However, the "1F1B" schedule inevitably leads to weight inconsistency and weight staleness issues due to the cross-training of different mini-batches across GPUs. To simultaneously address these two problems, in this paper, we propose an optimizer-dependent weight prediction strategy (a.k.a PipeOptim) for asynchronous pipeline training. The key insight of our proposal is that we employ a weight prediction strategy in the forward pass to ensure that each mini-batch uses consistent and staleness-free weights to compute the forward pass. To be concrete, we first construct the weight prediction scheme based on the update rule of the used optimizer when training the deep neural network models. Then throughout the "1F1B" pipelined training, each mini-batch is mandated to execute weight prediction ahead of the forward pass, subsequently employing the predicted weights to perform the forward pass. As a result, PipeOptim 1) inherits the advantage of the "1F1B" schedule and generates pretty high throughput, and 2) can ensure effective parameter learning regardless of the type of the used optimizer. To verify the effectiveness of our proposal, we conducted extensive experimental evaluations using eight different deep-learning models spanning three machine-learning tasks including image classification, sentiment analysis, and machine translation. The experiment results demonstrate that PipeOptim outperforms the popular pipelined approaches including GPipe, PipeDream, PipeDream-2BW, and SpecTrain. The code of PipeOptim can be accessible at https://github.com/guanleics/PipeOptim.
A Survey of Deep Visual Cross-Domain Few-Shot Learning
Mar 16, 2023
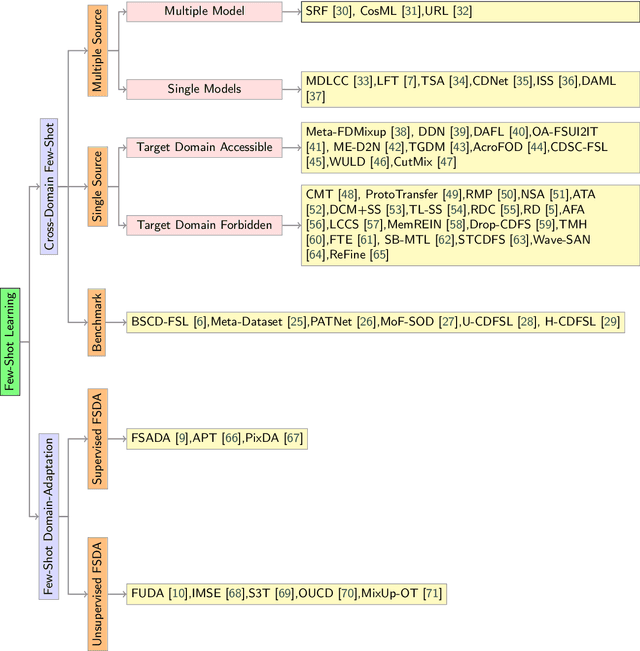
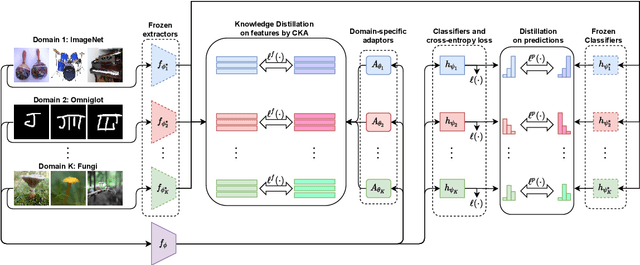
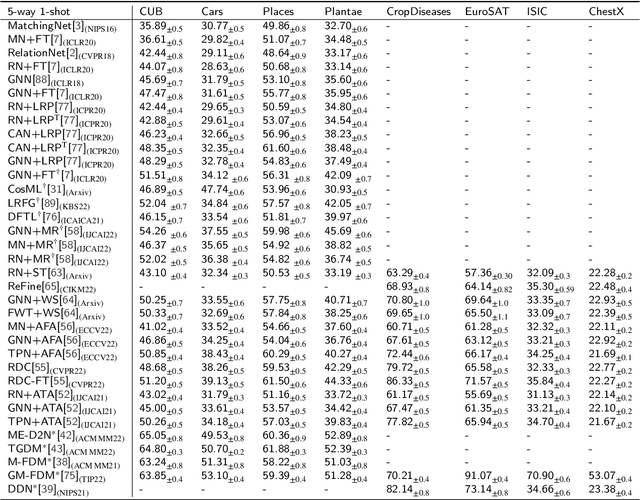
Abstract:Few-Shot transfer learning has become a major focus of research as it allows recognition of new classes with limited labeled data. While it is assumed that train and test data have the same data distribution, this is often not the case in real-world applications. This leads to decreased model transfer effects when the new class distribution differs significantly from the learned classes. Research into Cross-Domain Few-Shot (CDFS) has emerged to address this issue, forming a more challenging and realistic setting. In this survey, we provide a detailed taxonomy of CDFS from the problem setting and corresponding solutions view. We summarise the existing CDFS network architectures and discuss the solution ideas for each direction the taxonomy indicates. Furthermore, we introduce various CDFS downstream applications and outline classification, detection, and segmentation benchmarks and corresponding standards for evaluation. We also discuss the challenges of CDFS research and explore potential directions for future investigation. Through this review, we aim to provide comprehensive guidance on CDFS research, enabling researchers to gain insight into the state-of-the-art while allowing them to build upon existing solutions to develop their own CDFS models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge