Shuojue Yang
BCRNet: Enhancing Landmark Detection in Laparoscopic Liver Surgery via Bezier Curve Refinement
Jun 18, 2025


Abstract:Laparoscopic liver surgery, while minimally invasive, poses significant challenges in accurately identifying critical anatomical structures. Augmented reality (AR) systems, integrating MRI/CT with laparoscopic images based on 2D-3D registration, offer a promising solution for enhancing surgical navigation. A vital aspect of the registration progress is the precise detection of curvilinear anatomical landmarks in laparoscopic images. In this paper, we propose BCRNet (Bezier Curve Refinement Net), a novel framework that significantly enhances landmark detection in laparoscopic liver surgery primarily via the Bezier curve refinement strategy. The framework starts with a Multi-modal Feature Extraction (MFE) module designed to robustly capture semantic features. Then we propose Adaptive Curve Proposal Initialization (ACPI) to generate pixel-aligned Bezier curves and confidence scores for reliable initial proposals. Additionally, we design the Hierarchical Curve Refinement (HCR) mechanism to enhance these proposals iteratively through a multi-stage process, capturing fine-grained contextual details from multi-scale pixel-level features for precise Bezier curve adjustment. Extensive evaluations on the L3D and P2ILF datasets demonstrate that BCRNet outperforms state-of-the-art methods, achieving significant performance improvements. Code will be available.
ToolTipNet: A Segmentation-Driven Deep Learning Baseline for Surgical Instrument Tip Detection
Apr 13, 2025Abstract:In robot-assisted laparoscopic radical prostatectomy (RALP), the location of the instrument tip is important to register the ultrasound frame with the laparoscopic camera frame. A long-standing limitation is that the instrument tip position obtained from the da Vinci API is inaccurate and requires hand-eye calibration. Thus, directly computing the position of the tool tip in the camera frame using the vision-based method becomes an attractive solution. Besides, surgical instrument tip detection is the key component of other tasks, like surgical skill assessment and surgery automation. However, this task is challenging due to the small size of the tool tip and the articulation of the surgical instrument. Surgical instrument segmentation becomes relatively easy due to the emergence of the Segmentation Foundation Model, i.e., Segment Anything. Based on this advancement, we explore the deep learning-based surgical instrument tip detection approach that takes the part-level instrument segmentation mask as input. Comparison experiments with a hand-crafted image-processing approach demonstrate the superiority of the proposed method on simulated and real datasets.
Instrument-Splatting: Controllable Photorealistic Reconstruction of Surgical Instruments Using Gaussian Splatting
Mar 06, 2025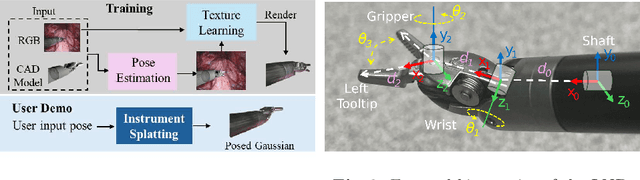
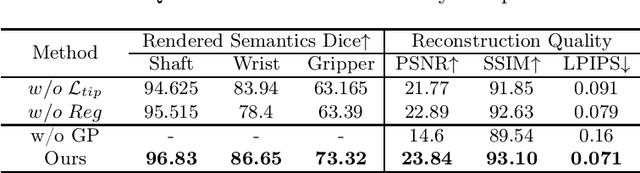
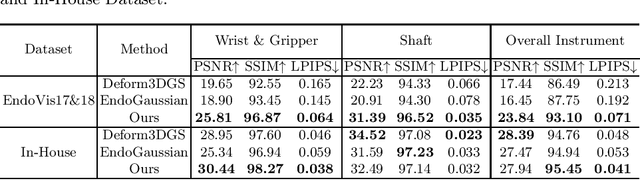
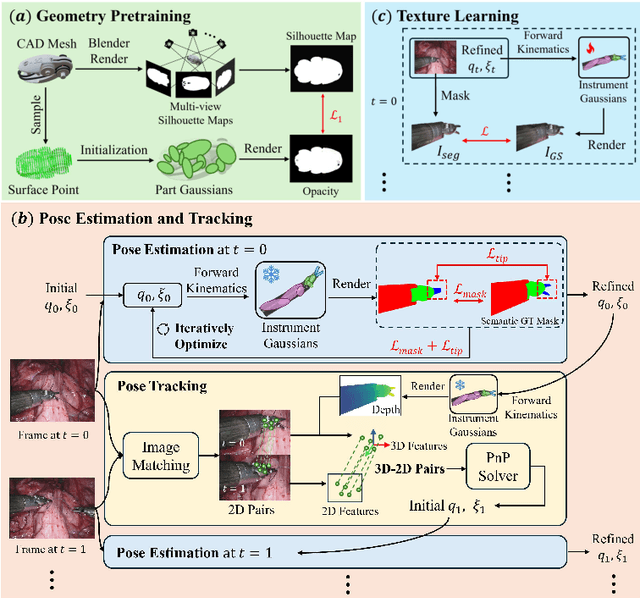
Abstract:Real2Sim is becoming increasingly important with the rapid development of surgical artificial intelligence (AI) and autonomy. In this work, we propose a novel Real2Sim methodology, \textit{Instrument-Splatting}, that leverages 3D Gaussian Splatting to provide fully controllable 3D reconstruction of surgical instruments from monocular surgical videos. To maintain both high visual fidelity and manipulability, we introduce a geometry pre-training to bind Gaussian point clouds on part mesh with accurate geometric priors and define a forward kinematics to control the Gaussians as flexible as real instruments. Afterward, to handle unposed videos, we design a novel instrument pose tracking method leveraging semantics-embedded Gaussians to robustly refine per-frame instrument poses and joint states in a render-and-compare manner, which allows our instrument Gaussian to accurately learn textures and reach photorealistic rendering. We validated our method on 2 publicly released surgical videos and 4 videos collected on ex vivo tissues and green screens. Quantitative and qualitative evaluations demonstrate the effectiveness and superiority of the proposed method.
Free-DyGS: Camera-Pose-Free Scene Reconstruction based on Gaussian Splatting for Dynamic Surgical Videos
Sep 02, 2024Abstract:Reconstructing endoscopic videos is crucial for high-fidelity visualization and the efficiency of surgical operations. Despite the importance, existing 3D reconstruction methods encounter several challenges, including stringent demands for accuracy, imprecise camera positioning, intricate dynamic scenes, and the necessity for rapid reconstruction. Addressing these issues, this paper presents the first camera-pose-free scene reconstruction framework, Free-DyGS, tailored for dynamic surgical videos, leveraging 3D Gaussian splatting technology. Our approach employs a frame-by-frame reconstruction strategy and is delineated into four distinct phases: Scene Initialization, Joint Learning, Scene Expansion, and Retrospective Learning. We introduce a Generalizable Gaussians Parameterization module within the Scene Initialization and Expansion phases to proficiently generate Gaussian attributes for each pixel from the RGBD frames. The Joint Learning phase is crafted to concurrently deduce scene deformation and camera pose, facilitated by an innovative flexible deformation module. In the scene expansion stage, the Gaussian points gradually grow as the camera moves. The Retrospective Learning phase is dedicated to enhancing the precision of scene deformation through the reassessment of prior frames. The efficacy of the proposed Free-DyGS is substantiated through experiments on two datasets: the StereoMIS and Hamlyn datasets. The experimental outcomes underscore that Free-DyGS surpasses conventional baseline models in both rendering fidelity and computational efficiency.
Deform3DGS: Flexible Deformation for Fast Surgical Scene Reconstruction with Gaussian Splatting
May 29, 2024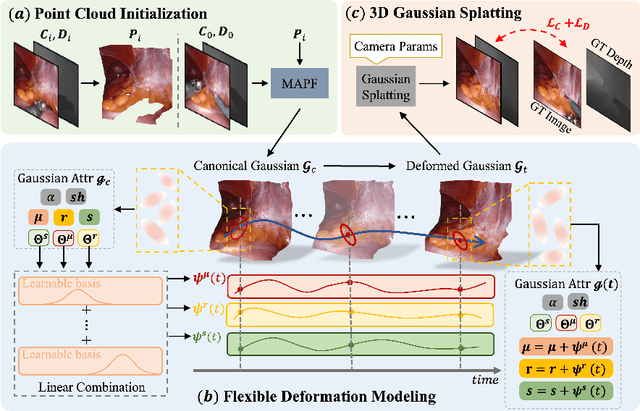
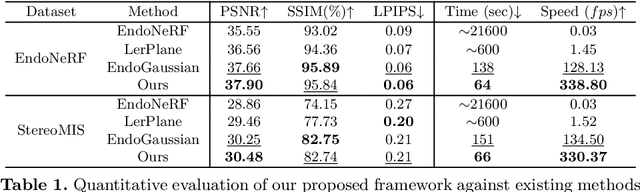

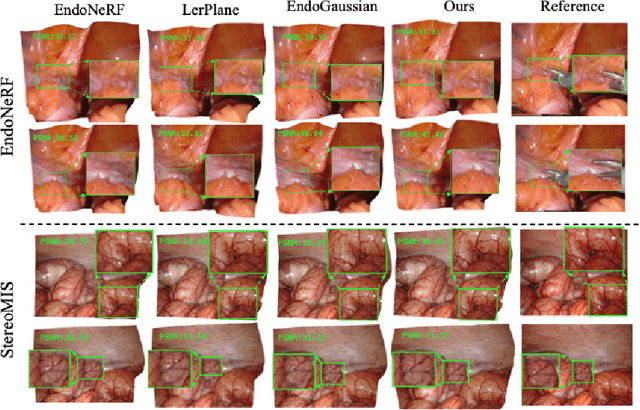
Abstract:Tissue deformation poses a key challenge for accurate surgical scene reconstruction. Despite yielding high reconstruction quality, existing methods suffer from slow rendering speeds and long training times, limiting their intraoperative applicability. Motivated by recent progress in 3D Gaussian Splatting, an emerging technology in real-time 3D rendering, this work presents a novel fast reconstruction framework, termed Deform3DGS, for deformable tissues during endoscopic surgery. Specifically, we introduce 3D GS into surgical scenes by integrating a point cloud initialization to improve reconstruction. Furthermore, we propose a novel flexible deformation modeling scheme (FDM) to learn tissue deformation dynamics at the level of individual Gaussians. Our FDM can model the surface deformation with efficient representations, allowing for real-time rendering performance. More importantly, FDM significantly accelerates surgical scene reconstruction, demonstrating considerable clinical values, particularly in intraoperative settings where time efficiency is crucial. Experiments on DaVinci robotic surgery videos indicate the efficacy of our approach, showcasing superior reconstruction fidelity PSNR: (37.90) and rendering speed (338.8 FPS) while substantially reducing training time to only 1 minute/scene.
Automatic Search for Photoacoustic Marker Using Automated Transrectal Ultrasound
Jul 20, 2023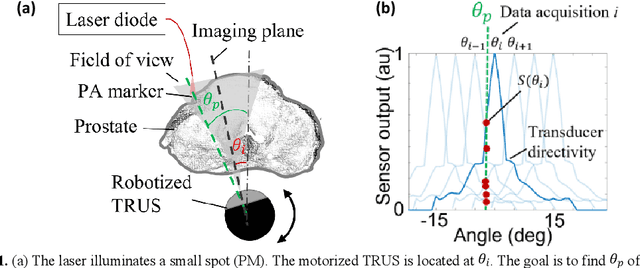
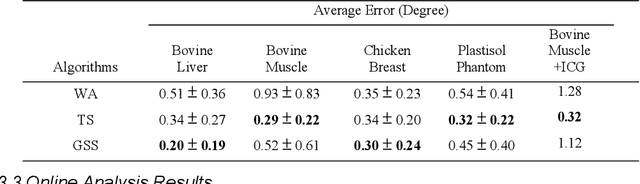
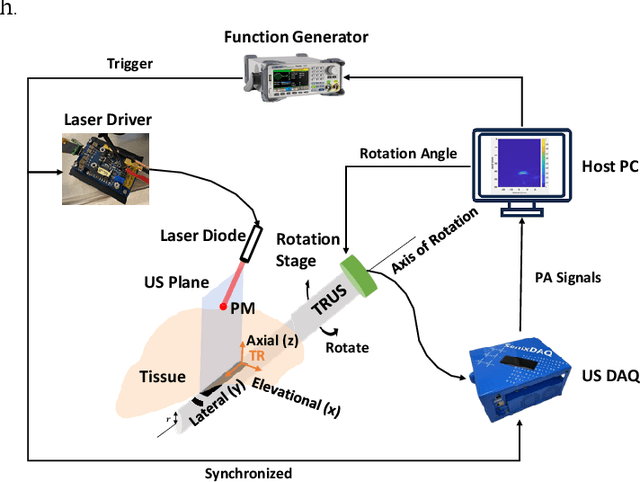
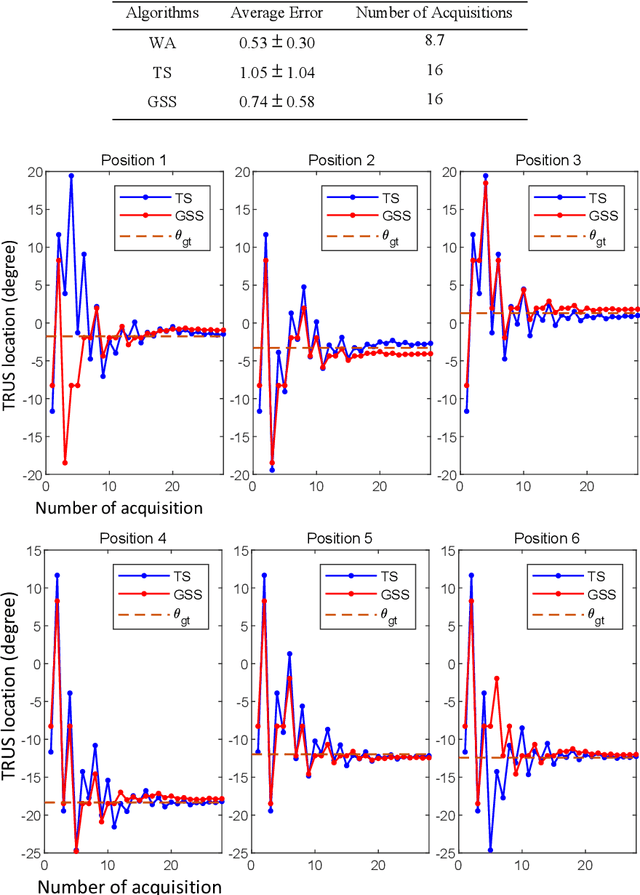
Abstract:Real-time transrectal ultrasound (TRUS) image guidance during robot-assisted laparoscopic radical prostatectomy has the potential to enhance surgery outcomes. Whether conventional or photoacoustic TRUS is used, the robotic system and the TRUS must be registered to each other. Accurate registration can be performed using photoacoustic (PA markers). However, this requires a manual search by an assistant [19]. This paper introduces the first automatic search for PA markers using a transrectal ultrasound robot. This effectively reduces the challenges associated with the da Vinci-TRUS registration. This paper investigated the performance of three search algorithms in simulation and experiment: Weighted Average (WA), Golden Section Search (GSS), and Ternary Search (TS). For validation, a surgical prostate scenario was mimicked and various ex vivo tissues were tested. As a result, the WA algorithm can achieve 0.53 degree average error after 9 data acquisitions, while the TS and GSS algorithm can achieve 0.29 degree and 0.48 degree average errors after 28 data acquisitions.
Arc-to-line frame registration method for ultrasound and photoacoustic image-guided intraoperative robot-assisted laparoscopic prostatectomy
Jun 21, 2023



Abstract:Purpose: To achieve effective robot-assisted laparoscopic prostatectomy, the integration of transrectal ultrasound (TRUS) imaging system which is the most widely used imaging modelity in prostate imaging is essential. However, manual manipulation of the ultrasound transducer during the procedure will significantly interfere with the surgery. Therefore, we propose an image co-registration algorithm based on a photoacoustic marker method, where the ultrasound / photoacoustic (US/PA) images can be registered to the endoscopic camera images to ultimately enable the TRUS transducer to automatically track the surgical instrument Methods: An optimization-based algorithm is proposed to co-register the images from the two different imaging modalities. The principles of light propagation and an uncertainty in PM detection were assumed in this algorithm to improve the stability and accuracy of the algorithm. The algorithm is validated using the previously developed US/PA image-guided system with a da Vinci surgical robot. Results: The target-registration-error (TRE) is measured to evaluate the proposed algorithm. In both simulation and experimental demonstration, the proposed algorithm achieved a sub-centimeter accuracy which is acceptable in practical clinics. The result is also comparable with our previous approach, and the proposed method can be implemented with a normal white light stereo camera and doesn't require highly accurate localization of the PM. Conclusion: The proposed frame registration algorithm enabled a simple yet efficient integration of commercial US/PA imaging system into laparoscopic surgical setting by leveraging the characteristic properties of acoustic wave propagation and laser excitation, contributing to automated US/PA image-guided surgical intervention applications.
PyMIC: A deep learning toolkit for annotation-efficient medical image segmentation
Aug 19, 2022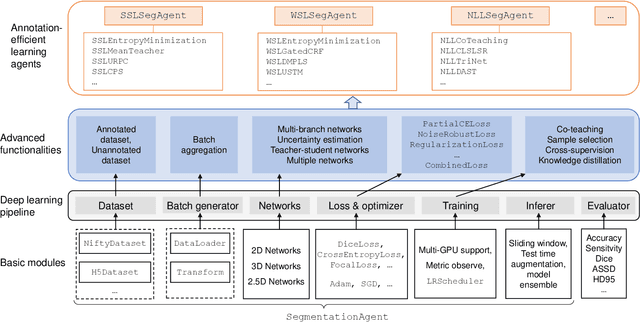
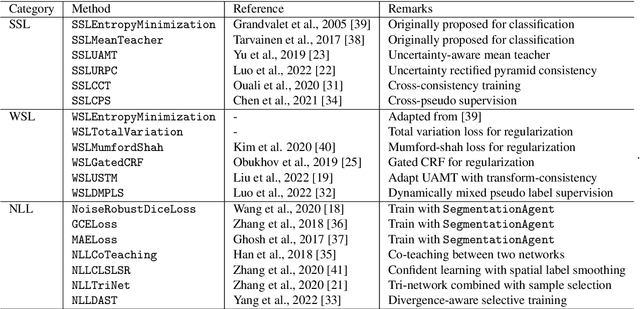
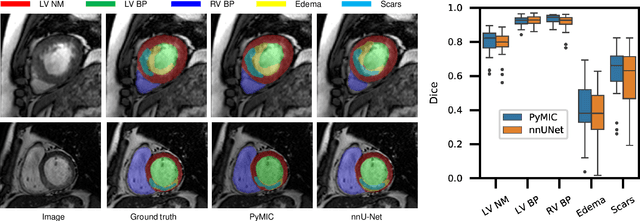
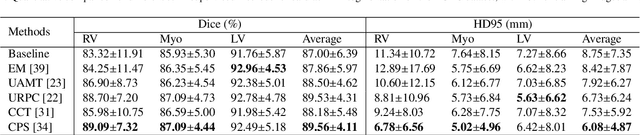
Abstract:Background and Objective: Existing deep learning platforms for medical image segmentation mainly focus on fully supervised segmentation that assumes full and accurate pixel-level annotations are available. We aim to develop a new deep learning toolkit to support annotation-efficient learning for medical image segmentation, which can accelerate and simply the development of deep learning models with limited annotation budget, e.g., learning from partial, sparse or noisy annotations. Methods: Our proposed toolkit named PyMIC is a modular deep learning platform for medical image segmentation tasks. In addition to basic components that support development of high-performance models for fully supervised segmentation, it contains several advanced components that are tailored for learning from imperfect annotations, such as loading annotated and unannounced images, loss functions for unannotated, partially or inaccurately annotated images, and training procedures for co-learning between multiple networks, etc. PyMIC is built on the PyTorch framework and supports development of semi-supervised, weakly supervised and noise-robust learning methods for medical image segmentation. Results: We present four illustrative medical image segmentation tasks based on PyMIC: (1) Achieving competitive performance on fully supervised learning; (2) Semi-supervised cardiac structure segmentation with only 10% training images annotated; (3) Weakly supervised segmentation using scribble annotations; and (4) Learning from noisy labels for chest radiograph segmentation. Conclusions: The PyMIC toolkit is easy to use and facilitates efficient development of medical image segmentation models with imperfect annotations. It is modular and flexible, which enables researchers to develop high-performance models with low annotation cost. The source code is available at: https://github.com/HiLab-git/PyMIC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge