Ke Zou
Generalist versus Specialist Vision Foundation Models for Ocular Disease and Oculomics
Sep 03, 2025Abstract:Medical foundation models, pre-trained with large-scale clinical data, demonstrate strong performance in diverse clinically relevant applications. RETFound, trained on nearly one million retinal images, exemplifies this approach in applications with retinal images. However, the emergence of increasingly powerful and multifold larger generalist foundation models such as DINOv2 and DINOv3 raises the question of whether domain-specific pre-training remains essential, and if so, what gap persists. To investigate this, we systematically evaluated the adaptability of DINOv2 and DINOv3 in retinal image applications, compared to two specialist RETFound models, RETFound-MAE and RETFound-DINOv2. We assessed performance on ocular disease detection and systemic disease prediction using two adaptation strategies: fine-tuning and linear probing. Data efficiency and adaptation efficiency were further analysed to characterise trade-offs between predictive performance and computational cost. Our results show that although scaling generalist models yields strong adaptability across diverse tasks, RETFound-DINOv2 consistently outperforms these generalist foundation models in ocular-disease detection and oculomics tasks, demonstrating stronger generalisability and data efficiency. These findings suggest that specialist retinal foundation models remain the most effective choice for clinical applications, while the narrowing gap with generalist foundation models suggests that continued data and model scaling can deliver domain-relevant gains and position them as strong foundations for future medical foundation models.
A Clinician-Friendly Platform for Ophthalmic Image Analysis Without Technical Barriers
Apr 22, 2025Abstract:Artificial intelligence (AI) shows remarkable potential in medical imaging diagnostics, but current models typically require retraining when deployed across different clinical centers, limiting their widespread adoption. We introduce GlobeReady, a clinician-friendly AI platform that enables ocular disease diagnosis without retraining/fine-tuning or technical expertise. GlobeReady achieves high accuracy across imaging modalities: 93.9-98.5% for an 11-category fundus photo dataset and 87.2-92.7% for a 15-category OCT dataset. Through training-free local feature augmentation, it addresses domain shifts across centers and populations, reaching an average accuracy of 88.9% across five centers in China, 86.3% in Vietnam, and 90.2% in the UK. The built-in confidence-quantifiable diagnostic approach further boosted accuracy to 94.9-99.4% (fundus) and 88.2-96.2% (OCT), while identifying out-of-distribution cases at 86.3% (49 CFP categories) and 90.6% (13 OCT categories). Clinicians from multiple countries rated GlobeReady highly (average 4.6 out of 5) for its usability and clinical relevance. These results demonstrate GlobeReady's robust, scalable diagnostic capability and potential to support ophthalmic care without technical barriers.
Benchmarking Next-Generation Reasoning-Focused Large Language Models in Ophthalmology: A Head-to-Head Evaluation on 5,888 Items
Apr 15, 2025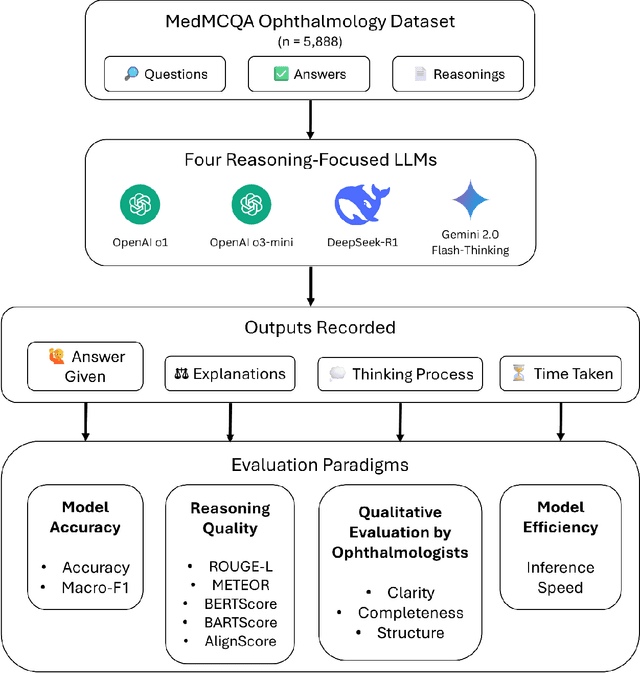
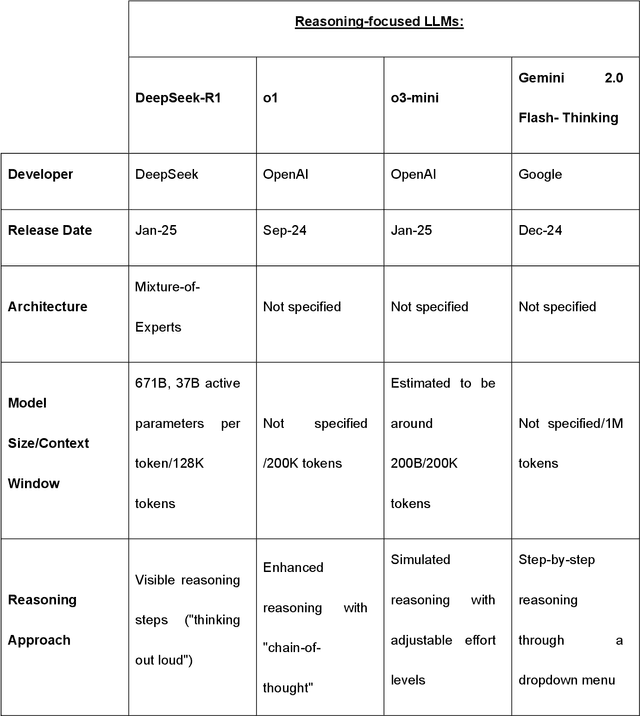
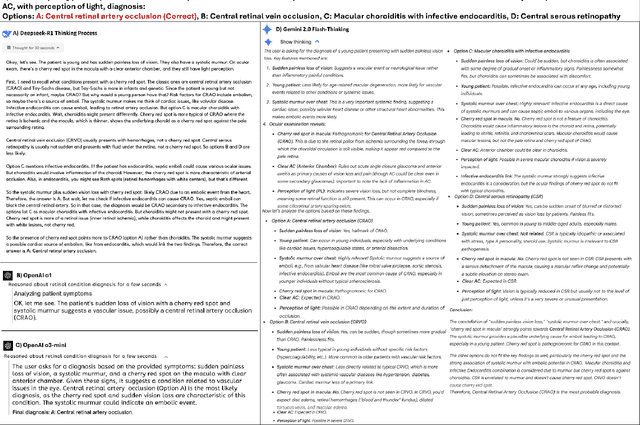
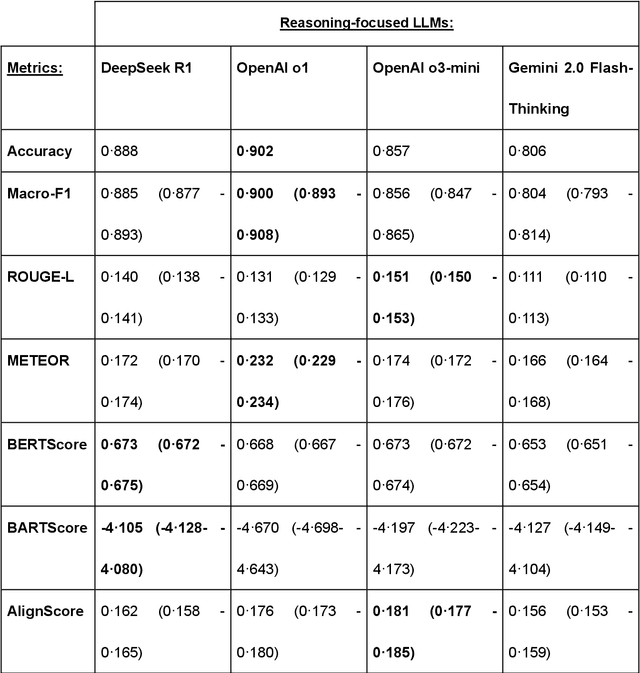
Abstract:Recent advances in reasoning-focused large language models (LLMs) mark a shift from general LLMs toward models designed for complex decision-making, a crucial aspect in medicine. However, their performance in specialized domains like ophthalmology remains underexplored. This study comprehensively evaluated and compared the accuracy and reasoning capabilities of four newly developed reasoning-focused LLMs, namely DeepSeek-R1, OpenAI o1, o3-mini, and Gemini 2.0 Flash-Thinking. Each model was assessed using 5,888 multiple-choice ophthalmology exam questions from the MedMCQA dataset in zero-shot setting. Quantitative evaluation included accuracy, Macro-F1, and five text-generation metrics (ROUGE-L, METEOR, BERTScore, BARTScore, and AlignScore), computed against ground-truth reasonings. Average inference time was recorded for a subset of 100 randomly selected questions. Additionally, two board-certified ophthalmologists qualitatively assessed clarity, completeness, and reasoning structure of responses to differential diagnosis questions.O1 (0.902) and DeepSeek-R1 (0.888) achieved the highest accuracy, with o1 also leading in Macro-F1 (0.900). The performance of models across the text-generation metrics varied: O3-mini excelled in ROUGE-L (0.151), o1 in METEOR (0.232), DeepSeek-R1 and o3-mini tied for BERTScore (0.673), DeepSeek-R1 (-4.105) and Gemini 2.0 Flash-Thinking (-4.127) performed best in BARTScore, while o3-mini (0.181) and o1 (0.176) led AlignScore. Inference time across the models varied, with DeepSeek-R1 being slowest (40.4 seconds) and Gemini 2.0 Flash-Thinking fastest (6.7 seconds). Qualitative evaluation revealed that DeepSeek-R1 and Gemini 2.0 Flash-Thinking tended to provide detailed and comprehensive intermediate reasoning, whereas o1 and o3-mini displayed concise and summarized justifications.
Vision-Amplified Semantic Entropy for Hallucination Detection in Medical Visual Question Answering
Mar 26, 2025Abstract:Multimodal large language models (MLLMs) have demonstrated significant potential in medical Visual Question Answering (VQA). Yet, they remain prone to hallucinations-incorrect responses that contradict input images, posing substantial risks in clinical decision-making. Detecting these hallucinations is essential for establishing trust in MLLMs among clinicians and patients, thereby enabling their real-world adoption. Current hallucination detection methods, especially semantic entropy (SE), have demonstrated promising hallucination detection capacity for LLMs. However, adapting SE to medical MLLMs by incorporating visual perturbations presents a dilemma. Weak perturbations preserve image content and ensure clinical validity, but may be overlooked by medical MLLMs, which tend to over rely on language priors. In contrast, strong perturbations can distort essential diagnostic features, compromising clinical interpretation. To address this issue, we propose Vision Amplified Semantic Entropy (VASE), which incorporates weak image transformations and amplifies the impact of visual input, to improve hallucination detection in medical VQA. We first estimate the semantic predictive distribution under weak visual transformations to preserve clinical validity, and then amplify visual influence by contrasting this distribution with that derived from a distorted image. The entropy of the resulting distribution is estimated as VASE. Experiments on two medical open-ended VQA datasets demonstrate that VASE consistently outperforms existing hallucination detection methods.
Is an Ultra Large Natural Image-Based Foundation Model Superior to a Retina-Specific Model for Detecting Ocular and Systemic Diseases?
Feb 10, 2025Abstract:The advent of foundation models (FMs) is transforming medical domain. In ophthalmology, RETFound, a retina-specific FM pre-trained sequentially on 1.4 million natural images and 1.6 million retinal images, has demonstrated high adaptability across clinical applications. Conversely, DINOv2, a general-purpose vision FM pre-trained on 142 million natural images, has shown promise in non-medical domains. However, its applicability to clinical tasks remains underexplored. To address this, we conducted head-to-head evaluations by fine-tuning RETFound and three DINOv2 models (large, base, small) for ocular disease detection and systemic disease prediction tasks, across eight standardized open-source ocular datasets, as well as the Moorfields AlzEye and the UK Biobank datasets. DINOv2-large model outperformed RETFound in detecting diabetic retinopathy (AUROC=0.850-0.952 vs 0.823-0.944, across three datasets, all P<=0.007) and multi-class eye diseases (AUROC=0.892 vs. 0.846, P<0.001). In glaucoma, DINOv2-base model outperformed RETFound (AUROC=0.958 vs 0.940, P<0.001). Conversely, RETFound achieved superior performance over all DINOv2 models in predicting heart failure, myocardial infarction, and ischaemic stroke (AUROC=0.732-0.796 vs 0.663-0.771, all P<0.001). These trends persisted even with 10% of the fine-tuning data. These findings showcase the distinct scenarios where general-purpose and domain-specific FMs excel, highlighting the importance of aligning FM selection with task-specific requirements to optimise clinical performance.
Are Traditional Deep Learning Model Approaches as Effective as a Retinal-Specific Foundation Model for Ocular and Systemic Disease Detection?
Jan 21, 2025Abstract:Background: RETFound, a self-supervised, retina-specific foundation model (FM), showed potential in downstream applications. However, its comparative performance with traditional deep learning (DL) models remains incompletely understood. This study aimed to evaluate RETFound against three ImageNet-pretrained supervised DL models (ResNet50, ViT-base, SwinV2) in detecting ocular and systemic diseases. Methods: We fine-tuned/trained RETFound and three DL models on full datasets, 50%, 20%, and fixed sample sizes (400, 200, 100 images, with half comprising disease cases; for each DR severity class, 100 and 50 cases were used. Fine-tuned models were tested internally using the SEED (53,090 images) and APTOS-2019 (3,672 images) datasets and externally validated on population-based (BES, CIEMS, SP2, UKBB) and open-source datasets (ODIR-5k, PAPILA, GAMMA, IDRiD, MESSIDOR-2). Model performance was compared using area under the receiver operating characteristic curve (AUC) and Z-tests with Bonferroni correction (P<0.05/3). Interpretation: Traditional DL models are mostly comparable to RETFound for ocular disease detection with large datasets. However, RETFound is superior in systemic disease detection with smaller datasets. These findings offer valuable insights into the respective merits and limitation of traditional models and FMs.
GEMeX: A Large-Scale, Groundable, and Explainable Medical VQA Benchmark for Chest X-ray Diagnosis
Nov 25, 2024



Abstract:Medical Visual Question Answering (VQA) is an essential technology that integrates computer vision and natural language processing to automatically respond to clinical inquiries about medical images. However, current medical VQA datasets exhibit two significant limitations: (1) they often lack visual and textual explanations for answers, which impedes their ability to satisfy the comprehension needs of patients and junior doctors; (2) they typically offer a narrow range of question formats, inadequately reflecting the diverse requirements encountered in clinical scenarios. These limitations pose significant challenges to the development of a reliable and user-friendly Med-VQA system. To address these challenges, we introduce a large-scale, Groundable, and Explainable Medical VQA benchmark for chest X-ray diagnosis (GEMeX), featuring several innovative components: (1) A multi-modal explainability mechanism that offers detailed visual and textual explanations for each question-answer pair, thereby enhancing answer comprehensibility; (2) Four distinct question types, open-ended, closed-ended, single-choice, and multiple-choice, that better reflect diverse clinical needs. We evaluated 10 representative large vision language models on GEMeX and found that they underperformed, highlighting the dataset's complexity. However, after fine-tuning a baseline model using the training set, we observed a significant performance improvement, demonstrating the dataset's effectiveness. The project is available at www.med-vqa.com/GEMeX.
LMOD: A Large Multimodal Ophthalmology Dataset and Benchmark for Large Vision-Language Models
Oct 02, 2024



Abstract:Ophthalmology relies heavily on detailed image analysis for diagnosis and treatment planning. While large vision-language models (LVLMs) have shown promise in understanding complex visual information, their performance on ophthalmology images remains underexplored. We introduce LMOD, a dataset and benchmark for evaluating LVLMs on ophthalmology images, covering anatomical understanding, diagnostic analysis, and demographic extraction. LMODincludes 21,993 images spanning optical coherence tomography, scanning laser ophthalmoscopy, eye photos, surgical scenes, and color fundus photographs. We benchmark 13 state-of-the-art LVLMs and find that they are far from perfect for comprehending ophthalmology images. Models struggle with diagnostic analysis and demographic extraction, reveal weaknesses in spatial reasoning, diagnostic analysis, handling out-of-domain queries, and safeguards for handling biomarkers of ophthalmology images.
MedSAM-U: Uncertainty-Guided Auto Multi-Prompt Adaptation for Reliable MedSAM
Sep 02, 2024



Abstract:The Medical Segment Anything Model (MedSAM) has shown remarkable performance in medical image segmentation, drawing significant attention in the field. However, its sensitivity to varying prompt types and locations poses challenges. This paper addresses these challenges by focusing on the development of reliable prompts that enhance MedSAM's accuracy. We introduce MedSAM-U, an uncertainty-guided framework designed to automatically refine multi-prompt inputs for more reliable and precise medical image segmentation. Specifically, we first train a Multi-Prompt Adapter integrated with MedSAM, creating MPA-MedSAM, to adapt to diverse multi-prompt inputs. We then employ uncertainty-guided multi-prompt to effectively estimate the uncertainties associated with the prompts and their initial segmentation results. In particular, a novel uncertainty-guided prompts adaptation technique is then applied automatically to derive reliable prompts and their corresponding segmentation outcomes. We validate MedSAM-U using datasets from multiple modalities to train a universal image segmentation model. Compared to MedSAM, experimental results on five distinct modal datasets demonstrate that the proposed MedSAM-U achieves an average performance improvement of 1.7\% to 20.5\% across uncertainty-guided prompts.
Enhancing Diagnostic Reliability of Foundation Model with Uncertainty Estimation in OCT Images
Jun 18, 2024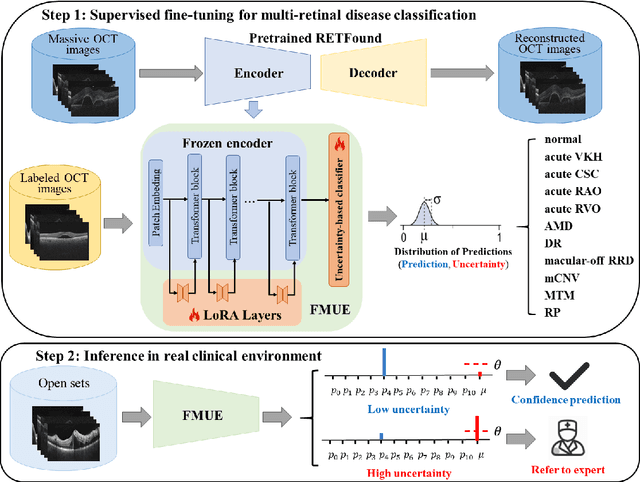
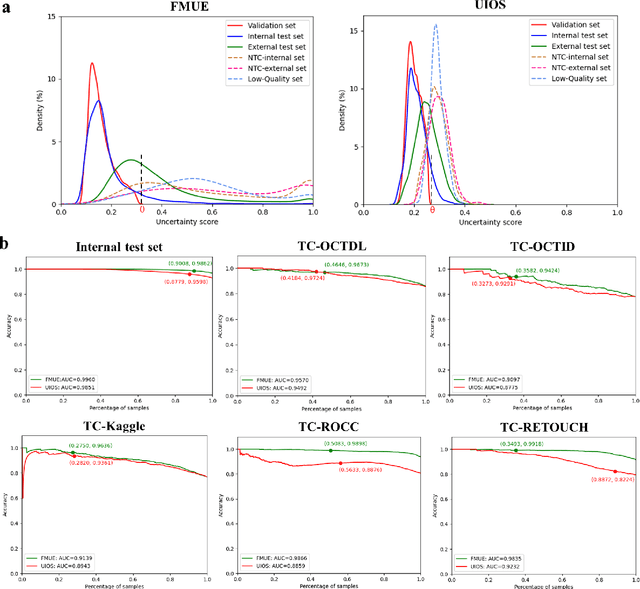
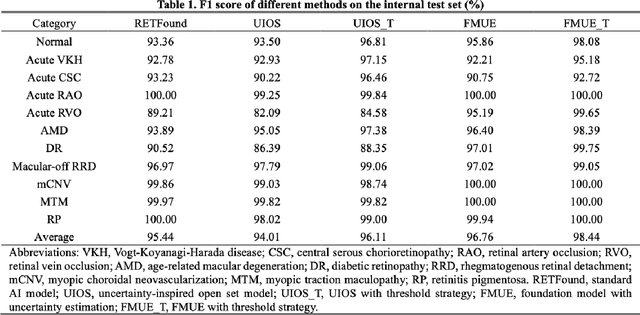
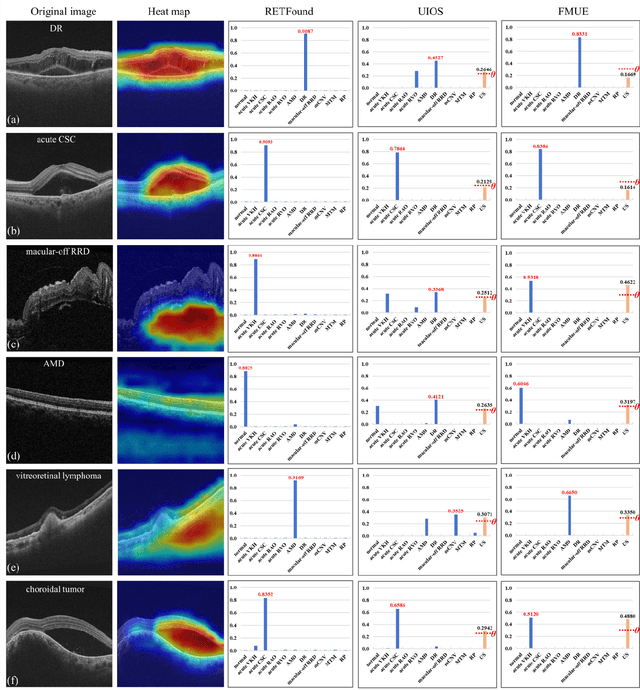
Abstract:Inability to express the confidence level and detect unseen classes has limited the clinical implementation of artificial intelligence in the real-world. We developed a foundation model with uncertainty estimation (FMUE) to detect 11 retinal conditions on optical coherence tomography (OCT). In the internal test set, FMUE achieved a higher F1 score of 96.76% than two state-of-the-art algorithms, RETFound and UIOS, and got further improvement with thresholding strategy to 98.44%. In the external test sets obtained from other OCT devices, FMUE achieved an accuracy of 88.75% and 92.73% before and after thresholding. Our model is superior to two ophthalmologists with a higher F1 score (95.17% vs. 61.93% &71.72%). Besides, our model correctly predicts high uncertainty scores for samples with ambiguous features, of non-target-category diseases, or with low-quality to prompt manual checks and prevent misdiagnosis. FMUE provides a trustworthy method for automatic retinal anomalies detection in the real-world clinical open set environment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge