Xiaojing Shen
DIFNet: Decentralized Information Filtering Fusion Neural Network with Unknown Correlation in Sensor Measurement Noises
Aug 26, 2025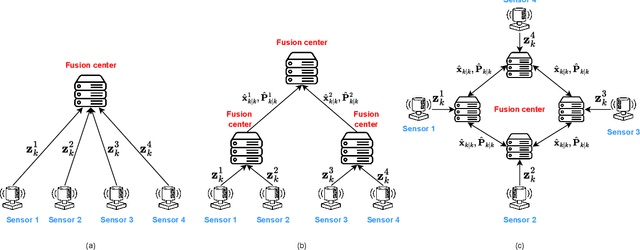
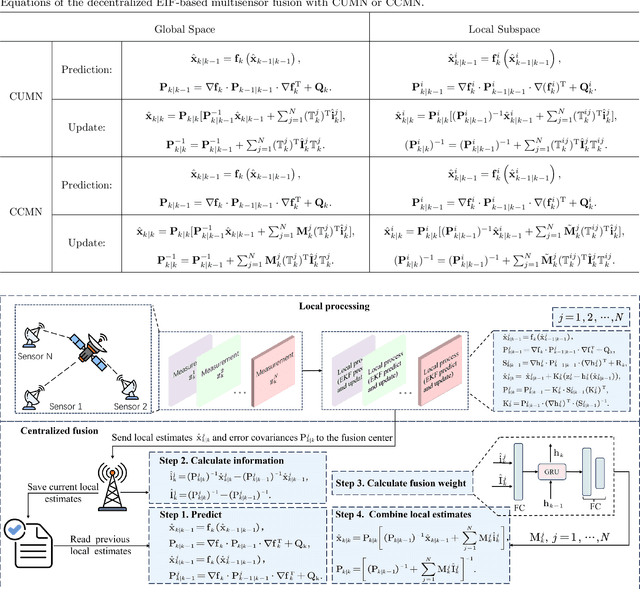
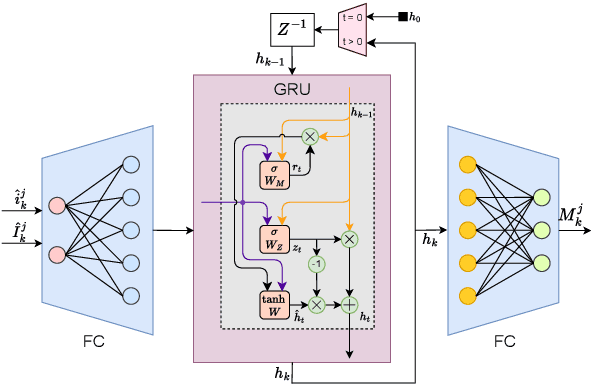
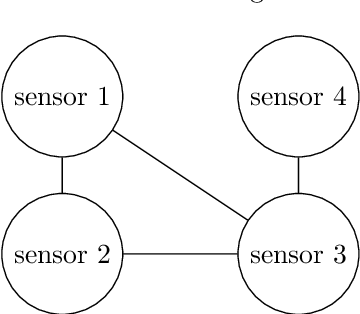
Abstract:In recent years, decentralized sensor networks have garnered significant attention in the field of state estimation owing to enhanced robustness, scalability, and fault tolerance. Optimal fusion performance can be achieved under fully connected communication and known noise correlation structures. To mitigate communication overhead, the global state estimation problem is decomposed into local subproblems through structured observation model. This ensures that even when the communication network is not fully connected, each sensor can achieve locally optimal estimates of its observable state components. To address the degradation of fusion accuracy induced by unknown correlations in measurement noise, this paper proposes a data-driven method, termed Decentralized Information Filter Neural Network (DIFNet), to learn unknown noise correlations in data for discrete-time nonlinear state space models with cross-correlated measurement noises. Numerical simulations demonstrate that DIFNet achieves superior fusion performance compared to conventional filtering methods and exhibits robust characteristics in more complex scenarios, such as the presence of time-varying noise. The source code used in our numerical experiment can be found online at https://wisdom-estimation.github.io/DIFNet_Demonstrate/.
A First-Order Algorithm for Graph Learning from Smooth Signals Under Partial Observability
Oct 08, 2024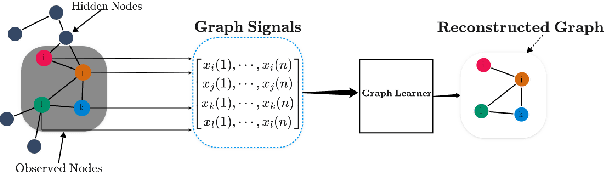

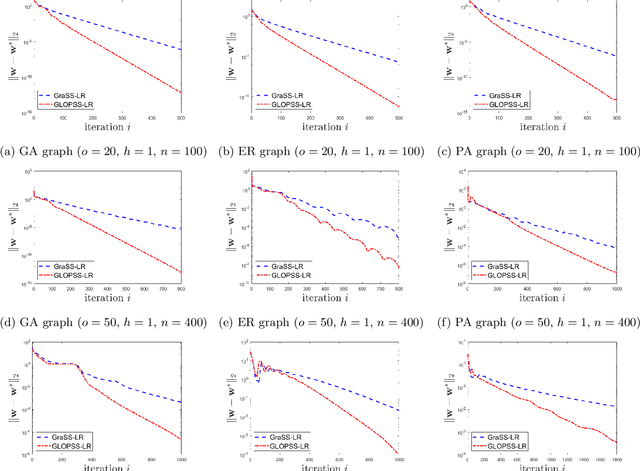
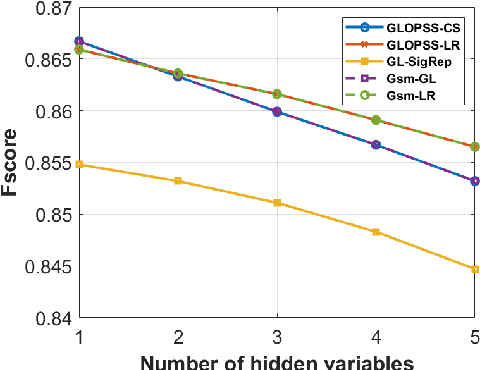
Abstract:Learning graph structures from smooth signals is a significant problem in data science and engineering. A common challenge in real-world scenarios is the availability of only partially observed nodes. While some studies have considered hidden nodes and proposed various optimization frameworks, existing methods often lack the practical efficiency needed for large-scale networks or fail to provide theoretical convergence guarantees. In this paper, we address the problem of inferring network topologies from smooth signals with partially observed nodes. We propose a first-order algorithmic framework that includes two variants: one based on column sparsity regularization and the other on a low-rank constraint. We establish theoretical convergence guarantees and demonstrate the linear convergence rate of our algorithms. Extensive experiments on both synthetic and real-world data show that our results align with theoretical predictions, exhibiting not only linear convergence but also superior speed compared to existing methods. To the best of our knowledge, this is the first work to propose a first-order algorithmic framework for inferring network structures from smooth signals under partial observability, offering both guaranteed linear convergence and practical effectiveness for large-scale networks.
PA-LLaVA: A Large Language-Vision Assistant for Human Pathology Image Understanding
Aug 18, 2024Abstract:The previous advancements in pathology image understanding primarily involved developing models tailored to specific tasks. Recent studies has demonstrated that the large vision-language model can enhance the performance of various downstream tasks in medical image understanding. In this study, we developed a domain-specific large language-vision assistant (PA-LLaVA) for pathology image understanding. Specifically, (1) we first construct a human pathology image-text dataset by cleaning the public medical image-text data for domain-specific alignment; (2) Using the proposed image-text data, we first train a pathology language-image pretraining (PLIP) model as the specialized visual encoder for pathology image, and then we developed scale-invariant connector to avoid the information loss caused by image scaling; (3) We adopt two-stage learning to train PA-LLaVA, first stage for domain alignment, and second stage for end to end visual question \& answering (VQA) task. In experiments, we evaluate our PA-LLaVA on both supervised and zero-shot VQA datasets, our model achieved the best overall performance among multimodal models of similar scale. The ablation experiments also confirmed the effectiveness of our design. We posit that our PA-LLaVA model and the datasets presented in this work can promote research in field of computational pathology. All codes are available at: https://github.com/ddw2AIGROUP2CQUPT/PA-LLaVA}{https://github.com/ddw2AIGROUP2CQUPT/PA-LLaVA
Confidence-aware multi-modality learning for eye disease screening
May 28, 2024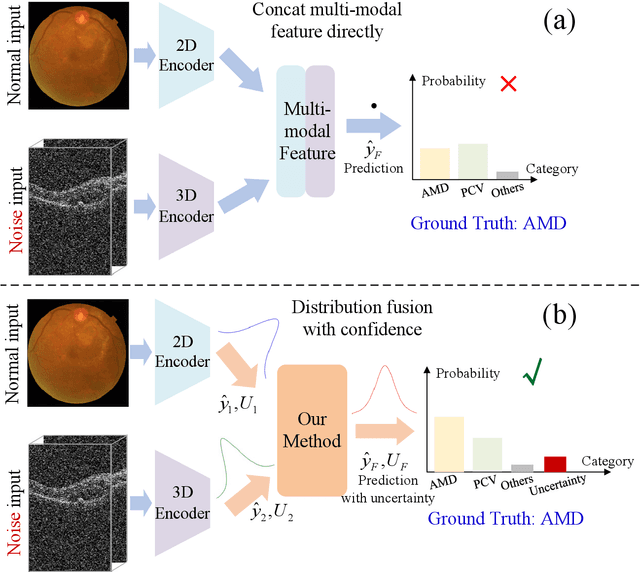
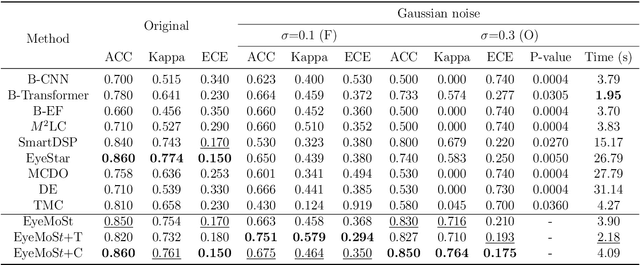
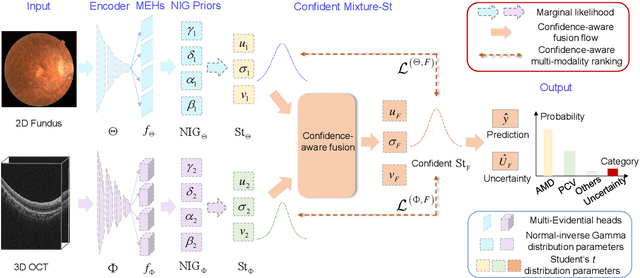
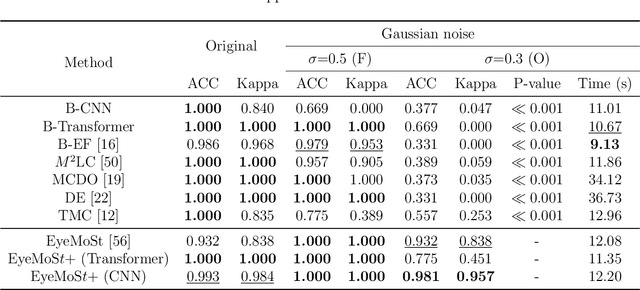
Abstract:Multi-modal ophthalmic image classification plays a key role in diagnosing eye diseases, as it integrates information from different sources to complement their respective performances. However, recent improvements have mainly focused on accuracy, often neglecting the importance of confidence and robustness in predictions for diverse modalities. In this study, we propose a novel multi-modality evidential fusion pipeline for eye disease screening. It provides a measure of confidence for each modality and elegantly integrates the multi-modality information using a multi-distribution fusion perspective. Specifically, our method first utilizes normal inverse gamma prior distributions over pre-trained models to learn both aleatoric and epistemic uncertainty for uni-modality. Then, the normal inverse gamma distribution is analyzed as the Student's t distribution. Furthermore, within a confidence-aware fusion framework, we propose a mixture of Student's t distributions to effectively integrate different modalities, imparting the model with heavy-tailed properties and enhancing its robustness and reliability. More importantly, the confidence-aware multi-modality ranking regularization term induces the model to more reasonably rank the noisy single-modal and fused-modal confidence, leading to improved reliability and accuracy. Experimental results on both public and internal datasets demonstrate that our model excels in robustness, particularly in challenging scenarios involving Gaussian noise and modality missing conditions. Moreover, our model exhibits strong generalization capabilities to out-of-distribution data, underscoring its potential as a promising solution for multimodal eye disease screening.
MedRG: Medical Report Grounding with Multi-modal Large Language Model
Apr 10, 2024



Abstract:Medical Report Grounding is pivotal in identifying the most relevant regions in medical images based on a given phrase query, a critical aspect in medical image analysis and radiological diagnosis. However, prevailing visual grounding approaches necessitate the manual extraction of key phrases from medical reports, imposing substantial burdens on both system efficiency and physicians. In this paper, we introduce a novel framework, Medical Report Grounding (MedRG), an end-to-end solution for utilizing a multi-modal Large Language Model to predict key phrase by incorporating a unique token, BOX, into the vocabulary to serve as an embedding for unlocking detection capabilities. Subsequently, the vision encoder-decoder jointly decodes the hidden embedding and the input medical image, generating the corresponding grounding box. The experimental results validate the effectiveness of MedRG, surpassing the performance of the existing state-of-the-art medical phrase grounding methods. This study represents a pioneering exploration of the medical report grounding task, marking the first-ever endeavor in this domain.
Destination-Constrained Linear Dynamical System Modeling in Set-Valued Frameworks
Mar 26, 2024
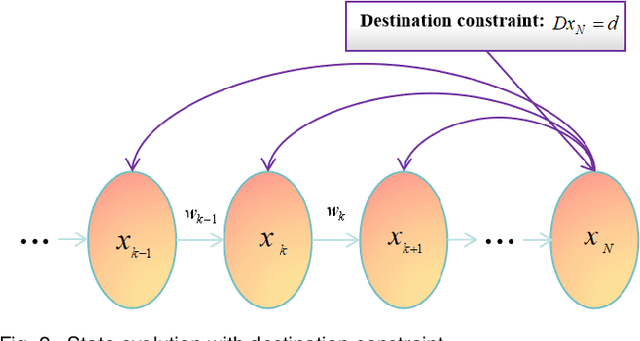
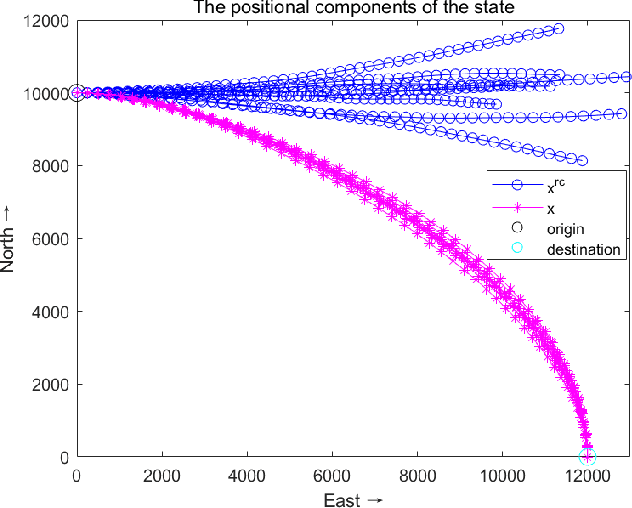
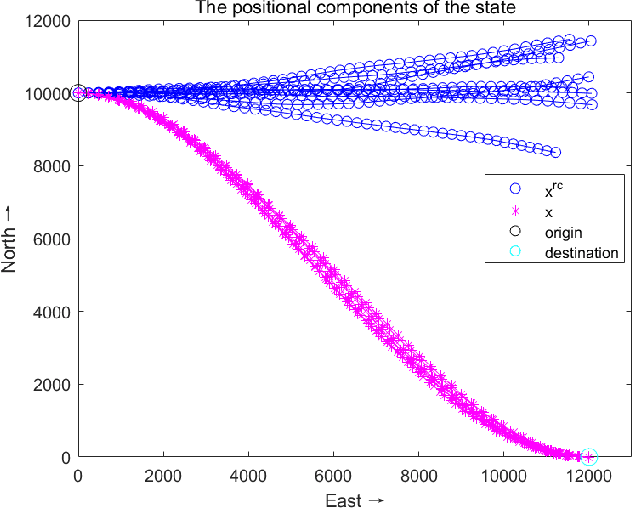
Abstract:Directional motion towards a specified destination is a common occurrence in physical processes and human societal activities. Utilizing this prior information can significantly improve the control and predictive performance of system models. This paper primarily focuses on reconstructing linear dynamic system models based on destination constraints in the set-valued framework. We treat destination constraints as inherent information in the state evolution process and employ convex optimization techniques to construct a coherent and robust state model. This refined model effectively captures the impact of destination constraints on the state evolution at each time step. Furthermore, we design an optimal weight matrix for the reconstructed model to ensure smoother and more natural trajectories of state evolution. We also analyze the theoretical guarantee of optimality for this weight matrix and the properties of the reconstructed model. Finally, simulation experiments verify that the reconstructed model has significant advantages over the unconstrained and unoptimized weighted models and constrains the evolution of state trajectories with different starting and ending points.
Uncertainty-informed Mutual Learning for Joint Medical Image Classification and Segmentation
Mar 30, 2023



Abstract:Classification and segmentation are crucial in medical image analysis as they enable accurate diagnosis and disease monitoring. However, current methods often prioritize the mutual learning features and shared model parameters, while neglecting the reliability of features and performances. In this paper, we propose a novel Uncertainty-informed Mutual Learning (UML) framework for reliable and interpretable medical image analysis. Our UML introduces reliability to joint classification and segmentation tasks, leveraging mutual learning with uncertainty to improve performance. To achieve this, we first use evidential deep learning to provide image-level and pixel-wise confidences. Then, an Uncertainty Navigator Decoder is constructed for better using mutual features and generating segmentation results. Besides, an Uncertainty Instructor is proposed to screen reliable masks for classification. Overall, UML could produce confidence estimation in features and performance for each link (classification and segmentation). The experiments on the public datasets demonstrate that our UML outperforms existing methods in terms of both accuracy and robustness. Our UML has the potential to explore the development of more reliable and explainable medical image analysis models. We will release the codes for reproduction after acceptance.
Reliable Multimodality Eye Disease Screening via Mixture of Student's t Distributions
Mar 17, 2023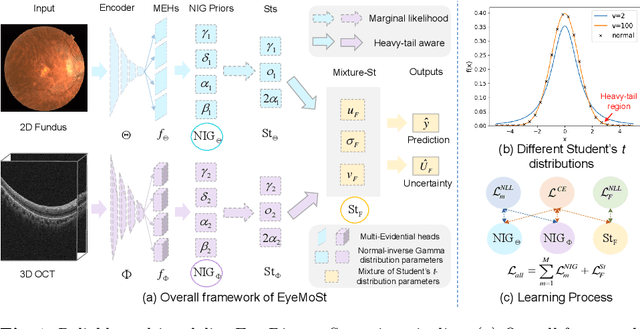
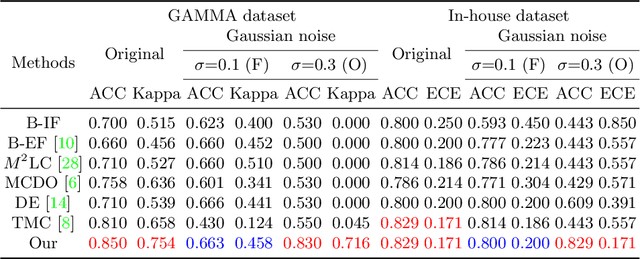
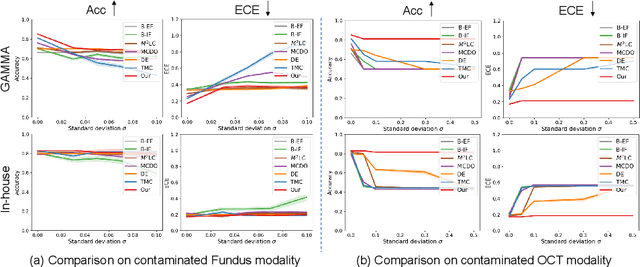

Abstract:Multimodality eye disease screening is crucial in ophthalmology as it integrates information from diverse sources to complement their respective performances. However, the existing methods are weak in assessing the reliability of each unimodality, and directly fusing an unreliable modality may cause screening errors. To address this issue, we introduce a novel multimodality evidential fusion pipeline for eye disease screening, EyeMoS$t$, which provides a measure of confidence for unimodality and elegantly integrates the multimodality information from a multi-distribution fusion perspective. Specifically, our model estimates both local uncertainty for unimodality and global uncertainty for the fusion modality to produce reliable classification results. More importantly, the proposed mixture of Student's $t$ distributions adaptively integrates different modalities to endow the model with heavy-tailed properties, increasing robustness and reliability. Our experimental findings on both public and in-house datasets show that our model is more reliable than current methods. Additionally, EyeMos$t$ has the potential ability to serve as a data quality discriminator, enabling reliable decision-making for multimodality eye disease screening.
A Review of Uncertainty Estimation and its Application in Medical Imaging
Feb 16, 2023



Abstract:The use of AI systems in healthcare for the early screening of diseases is of great clinical importance. Deep learning has shown great promise in medical imaging, but the reliability and trustworthiness of AI systems limit their deployment in real clinical scenes, where patient safety is at stake. Uncertainty estimation plays a pivotal role in producing a confidence evaluation along with the prediction of the deep model. This is particularly important in medical imaging, where the uncertainty in the model's predictions can be used to identify areas of concern or to provide additional information to the clinician. In this paper, we review the various types of uncertainty in deep learning, including aleatoric uncertainty, epistemic uncertainty, and out-of-distribution uncertainty, and we discuss how they can be estimated in medical imaging. We also review recent advances in deep learning models that incorporate uncertainty estimation in medical imaging. Finally, we discuss the challenges and future directions in uncertainty estimation in deep learning for medical imaging. We hope this review will ignite further interest in the community and provide researchers with an up-to-date reference regarding applications of uncertainty estimation models in medical imaging.
EvidenceCap: Towards trustworthy medical image segmentation via evidential identity cap
Jan 01, 2023



Abstract:Medical image segmentation (MIS) is essential for supporting disease diagnosis and treatment effect assessment. Despite considerable advances in artificial intelligence (AI) for MIS, clinicians remain skeptical of its utility, maintaining low confidence in such black box systems, with this problem being exacerbated by low generalization for out-of-distribution (OOD) data. To move towards effective clinical utilization, we propose a foundation model named EvidenceCap, which makes the box transparent in a quantifiable way by uncertainty estimation. EvidenceCap not only makes AI visible in regions of uncertainty and OOD data, but also enhances the reliability, robustness, and computational efficiency of MIS. Uncertainty is modeled explicitly through subjective logic theory to gather strong evidence from features. We show the effectiveness of EvidenceCap in three segmentation datasets and apply it to the clinic. Our work sheds light on clinical safe applications and explainable AI, and can contribute towards trustworthiness in the medical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge