Mengkang Lu
PAINT: Pathology-Aware Integrated Next-Scale Transformation for Virtual Immunohistochemistry
Jan 22, 2026Abstract:Virtual immunohistochemistry (IHC) aims to computationally synthesize molecular staining patterns from routine Hematoxylin and Eosin (H\&E) images, offering a cost-effective and tissue-efficient alternative to traditional physical staining. However, this task is particularly challenging: H\&E morphology provides ambiguous cues about protein expression, and similar tissue structures may correspond to distinct molecular states. Most existing methods focus on direct appearance synthesis to implicitly achieve cross-modal generation, often resulting in semantic inconsistencies due to insufficient structural priors. In this paper, we propose Pathology-Aware Integrated Next-Scale Transformation (PAINT), a visual autoregressive framework that reformulates the synthesis process as a structure-first conditional generation task. Unlike direct image translation, PAINT enforces a causal order by resolving molecular details conditioned on a global structural layout. Central to this approach is the introduction of a Spatial Structural Start Map (3S-Map), which grounds the autoregressive initialization in observed morphology, ensuring deterministic, spatially aligned synthesis. Experiments on the IHC4BC and MIST datasets demonstrate that PAINT outperforms state-of-the-art methods in structural fidelity and clinical downstream tasks, validating the potential of structure-guided autoregressive modeling.
Deformable Medical Image Registration with Effective Anatomical Structure Representation and Divide-and-Conquer Network
Jun 24, 2025Abstract:Effective representation of Regions of Interest (ROI) and independent alignment of these ROIs can significantly enhance the performance of deformable medical image registration (DMIR). However, current learning-based DMIR methods have limitations. Unsupervised techniques disregard ROI representation and proceed directly with aligning pairs of images, while weakly-supervised methods heavily depend on label constraints to facilitate registration. To address these issues, we introduce a novel ROI-based registration approach named EASR-DCN. Our method represents medical images through effective ROIs and achieves independent alignment of these ROIs without requiring labels. Specifically, we first used a Gaussian mixture model for intensity analysis to represent images using multiple effective ROIs with distinct intensities. Furthermore, we propose a novel Divide-and-Conquer Network (DCN) to process these ROIs through separate channels to learn feature alignments for each ROI. The resultant correspondences are seamlessly integrated to generate a comprehensive displacement vector field. Extensive experiments were performed on three MRI and one CT datasets to showcase the superior accuracy and deformation reduction efficacy of our EASR-DCN. Compared to VoxelMorph, our EASR-DCN achieved improvements of 10.31\% in the Dice score for brain MRI, 13.01\% for cardiac MRI, and 5.75\% for hippocampus MRI, highlighting its promising potential for clinical applications. The code for this work will be released upon acceptance of the paper.
Each Test Image Deserves A Specific Prompt: Continual Test-Time Adaptation for 2D Medical Image Segmentation
Nov 30, 2023Abstract:Distribution shift widely exists in medical images acquired from different medical centres and poses a significant obstacle to deploying the pre-trained semantic segmentation model in real-world applications. Test-time adaptation has proven its effectiveness in tackling the cross-domain distribution shift during inference. However, most existing methods achieve adaptation by updating the pre-trained models, rendering them susceptible to error accumulation and catastrophic forgetting when encountering a series of distribution shifts (i.e., under the continual test-time adaptation setup). To overcome these challenges caused by updating the models, in this paper, we freeze the pre-trained model and propose the Visual Prompt-based Test-Time Adaptation (VPTTA) method to train a specific prompt for each test image to align the statistics in the batch normalization layers. Specifically, we present the low-frequency prompt, which is lightweight with only a few parameters and can be effectively trained in a single iteration. To enhance prompt initialization, we equip VPTTA with a memory bank to benefit the current prompt from previous ones. Additionally, we design a warm-up mechanism, which mixes source and target statistics to construct warm-up statistics, thereby facilitating the training process. Extensive experiments demonstrate the superiority of our VPTTA over other state-of-the-art methods on two medical image segmentation benchmark tasks. The code and weights of pre-trained source models are available at https://github.com/Chen-Ziyang/VPTTA.
Segment Together: A Versatile Paradigm for Semi-Supervised Medical Image Segmentation
Nov 20, 2023



Abstract:Annotation scarcity has become a major obstacle for training powerful deep-learning models for medical image segmentation, restricting their deployment in clinical scenarios. To address it, semi-supervised learning by exploiting abundant unlabeled data is highly desirable to boost the model training. However, most existing works still focus on limited medical tasks and underestimate the potential of learning across diverse tasks and multiple datasets. Therefore, in this paper, we introduce a \textbf{Ver}satile \textbf{Semi}-supervised framework (VerSemi) to point out a new perspective that integrates various tasks into a unified model with a broad label space, to exploit more unlabeled data for semi-supervised medical image segmentation. Specifically, we introduce a dynamic task-prompted design to segment various targets from different datasets. Next, this unified model is used to identify the foreground regions from all labeled data, to capture cross-dataset semantics. Particularly, we create a synthetic task with a cutmix strategy to augment foreground targets within the expanded label space. To effectively utilize unlabeled data, we introduce a consistency constraint. This involves aligning aggregated predictions from various tasks with those from the synthetic task, further guiding the model in accurately segmenting foreground regions during training. We evaluated our VerSemi model on four public benchmarking datasets. Extensive experiments demonstrated that VerSemi can consistently outperform the second-best method by a large margin (e.g., an average 2.69\% Dice gain on four datasets), setting new SOTA performance for semi-supervised medical image segmentation. The code will be released.
Discrepancy Matters: Learning from Inconsistent Decoder Features for Consistent Semi-supervised Medical Image Segmentation
Sep 26, 2023Abstract:Semi-supervised learning (SSL) has been proven beneficial for mitigating the issue of limited labeled data especially on the task of volumetric medical image segmentation. Unlike previous SSL methods which focus on exploring highly confident pseudo-labels or developing consistency regularization schemes, our empirical findings suggest that inconsistent decoder features emerge naturally when two decoders strive to generate consistent predictions. Based on the observation, we first analyze the treasure of discrepancy in learning towards consistency, under both pseudo-labeling and consistency regularization settings, and subsequently propose a novel SSL method called LeFeD, which learns the feature-level discrepancy obtained from two decoders, by feeding the discrepancy as a feedback signal to the encoder. The core design of LeFeD is to enlarge the difference by training differentiated decoders, and then learn from the inconsistent information iteratively. We evaluate LeFeD against eight state-of-the-art (SOTA) methods on three public datasets. Experiments show LeFeD surpasses competitors without any bells and whistles such as uncertainty estimation and strong constraints, as well as setting a new state-of-the-art for semi-supervised medical image segmentation. Code is available at \textcolor{cyan}{https://github.com/maxwell0027/LeFeD}
A Novel Unified Conditional Score-based Generative Framework for Multi-modal Medical Image Completion
Jul 07, 2022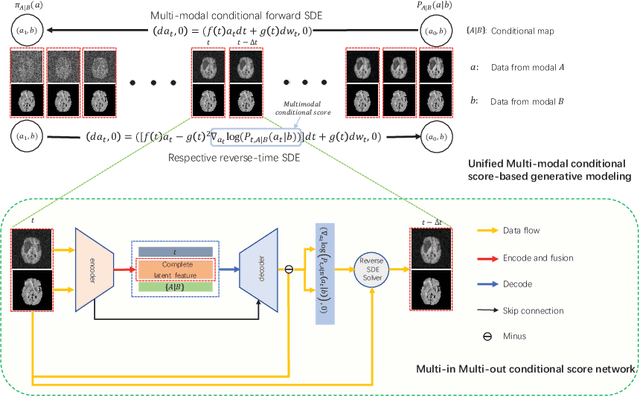
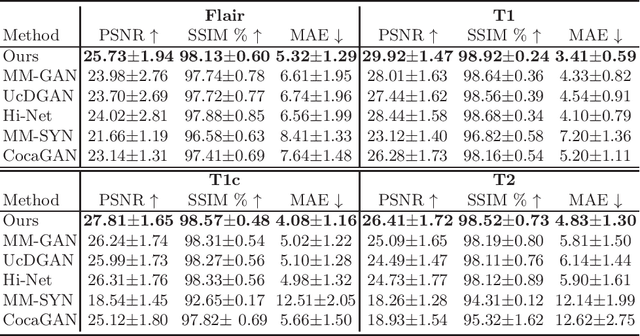
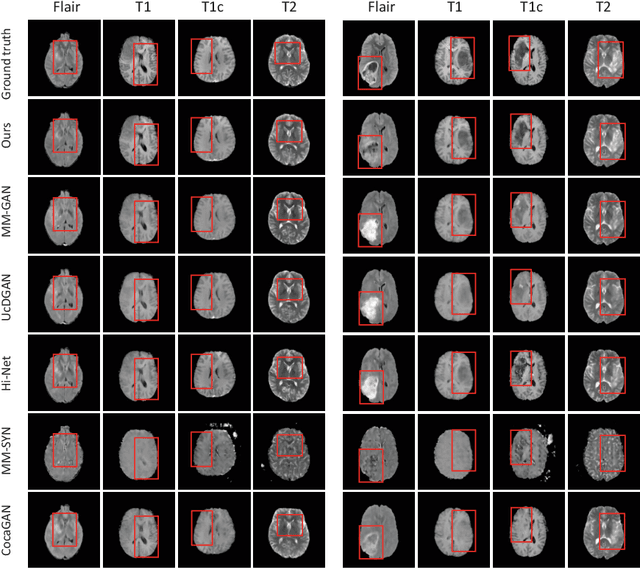
Abstract:Multi-modal medical image completion has been extensively applied to alleviate the missing modality issue in a wealth of multi-modal diagnostic tasks. However, for most existing synthesis methods, their inferences of missing modalities can collapse into a deterministic mapping from the available ones, ignoring the uncertainties inherent in the cross-modal relationships. Here, we propose the Unified Multi-Modal Conditional Score-based Generative Model (UMM-CSGM) to take advantage of Score-based Generative Model (SGM) in modeling and stochastically sampling a target probability distribution, and further extend SGM to cross-modal conditional synthesis for various missing-modality configurations in a unified framework. Specifically, UMM-CSGM employs a novel multi-in multi-out Conditional Score Network (mm-CSN) to learn a comprehensive set of cross-modal conditional distributions via conditional diffusion and reverse generation in the complete modality space. In this way, the generation process can be accurately conditioned by all available information, and can fit all possible configurations of missing modalities in a single network. Experiments on BraTS19 dataset show that the UMM-CSGM can more reliably synthesize the heterogeneous enhancement and irregular area in tumor-induced lesions for any missing modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge