Nizhuan Wang
Linguistics and Human Brain: A Perspective of Computational Neuroscience
Feb 09, 2026Abstract:Elucidating the language-brain relationship requires bridging the methodological gap between the abstract theoretical frameworks of linguistics and the empirical neural data of neuroscience. Serving as an interdisciplinary cornerstone, computational neuroscience formalizes the hierarchical and dynamic structures of language into testable neural models through modeling, simulation, and data analysis. This enables a computational dialogue between linguistic hypotheses and neural mechanisms. Recent advances in deep learning, particularly large language models (LLMs), have powerfully advanced this pursuit. Their high-dimensional representational spaces provide a novel scale for exploring the neural basis of linguistic processing, while the "model-brain alignment" framework offers a methodology to evaluate the biological plausibility of language-related theories.
Prototype-Based Disentanglement for Controllable Dysarthric Speech Synthesis
Feb 09, 2026Abstract:Dysarthric speech exhibits high variability and limited labeled data, posing major challenges for both automatic speech recognition (ASR) and assistive speech technologies. Existing approaches rely on synthetic data augmentation or speech reconstruction, yet often entangle speaker identity with pathological articulation, limiting controllability and robustness. In this paper, we propose ProtoDisent-TTS, a prototype-based disentanglement TTS framework built on a pre-trained text-to-speech backbone that factorizes speaker timbre and dysarthric articulation within a unified latent space. A pathology prototype codebook provides interpretable and controllable representations of healthy and dysarthric speech patterns, while a dual-classifier objective with a gradient reversal layer enforces invariance of speaker embeddings to pathological attributes. Experiments on the TORGO dataset demonstrate that this design enables bidirectional transformation between healthy and dysarthric speech, leading to consistent ASR performance gains and robust, speaker-aware speech reconstruction.
Region-aware Spatiotemporal Modeling with Collaborative Domain Generalization for Cross-Subject EEG Emotion Recognition
Jan 22, 2026Abstract:Cross-subject EEG-based emotion recognition (EER) remains challenging due to strong inter-subject variability, which induces substantial distribution shifts in EEG signals, as well as the high complexity of emotion-related neural representations in both spatial organization and temporal evolution. Existing approaches typically improve spatial modeling, temporal modeling, or generalization strategies in isolation, which limits their ability to align representations across subjects while capturing multi-scale dynamics and suppressing subject-specific bias within a unified framework. To address these gaps, we propose a Region-aware Spatiotemporal Modeling framework with Collaborative Domain Generalization (RSM-CoDG) for cross-subject EEG emotion recognition. RSM-CoDG incorporates neuroscience priors derived from functional brain region partitioning to construct region-level spatial representations, thereby improving cross-subject comparability. It also employs multi-scale temporal modeling to characterize the dynamic evolution of emotion-evoked neural activity. In addition, the framework employs a collaborative domain generalization strategy, incorporating multidimensional constraints to reduce subject-specific bias in a fully unseen target subject setting, which enhances the generalization to unknown individuals. Extensive experimental results on SEED series datasets demonstrate that RSM-CoDG consistently outperforms existing competing methods, providing an effective approach for improving robustness. The source code is available at https://github.com/RyanLi-X/RSM-CoDG.
LEREL: Lipschitz Continuity-Constrained Emotion Recognition Ensemble Learning For Electroencephalography
Apr 12, 2025



Abstract:Accurate and efficient perception of emotional states in oneself and others is crucial, as emotion-related disorders are associated with severe psychosocial impairments. While electroencephalography (EEG) offers a powerful tool for emotion detection, current EEG-based emotion recognition (EER) methods face key limitations: insufficient model stability, limited accuracy in processing high-dimensional nonlinear EEG signals, and poor robustness against intra-subject variability and signal noise. To address these challenges, we propose LEREL (Lipschitz continuity-constrained Emotion Recognition Ensemble Learning), a novel framework that significantly enhances both the accuracy and robustness of emotion recognition performance. The LEREL framework employs Lipschitz continuity constraints to enhance model stability and generalization in EEG emotion recognition, reducing signal variability and noise susceptibility while maintaining strong performance on small-sample datasets. The ensemble learning strategy reduces single-model bias and variance through multi-classifier decision fusion, further optimizing overall performance. Experimental results on three public benchmark datasets (EAV, FACED and SEED) demonstrate LEREL's effectiveness, achieving average recognition accuracies of 76.43%, 83.00% and 89.22%, respectively.
Information Bottleneck-Guided Heterogeneous Graph Learning for Interpretable Neurodevelopmental Disorder Diagnosis
Feb 28, 2025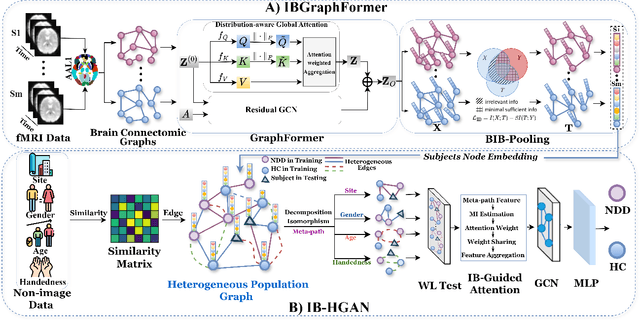
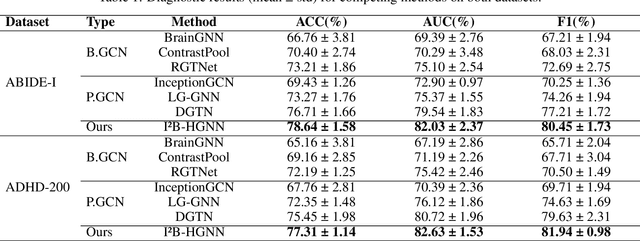
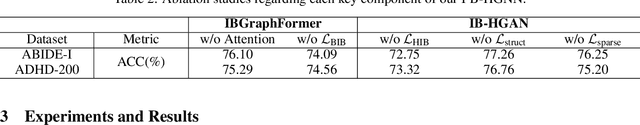
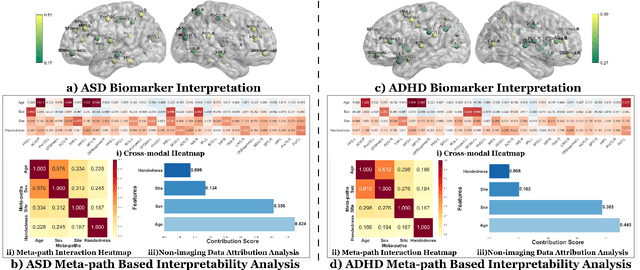
Abstract:Developing interpretable models for diagnosing neurodevelopmental disorders (NDDs) is highly valuable yet challenging, primarily due to the complexity of encoding, decoding and integrating imaging and non-imaging data. Many existing machine learning models struggle to provide comprehensive interpretability, often failing to extract meaningful biomarkers from imaging data, such as functional magnetic resonance imaging (fMRI), or lacking mechanisms to explain the significance of non-imaging data. In this paper, we propose the Interpretable Information Bottleneck Heterogeneous Graph Neural Network (I2B-HGNN), a novel framework designed to learn from fine-grained local patterns to comprehensive global multi-modal interactions. This framework comprises two key modules. The first module, the Information Bottleneck Graph Transformer (IBGraphFormer) for local patterns, integrates global modeling with brain connectomic-constrained graph neural networks to identify biomarkers through information bottleneck-guided pooling. The second module, the Information Bottleneck Heterogeneous Graph Attention Network (IB-HGAN) for global multi-modal interactions, facilitates interpretable multi-modal fusion of imaging and non-imaging data using heterogeneous graph neural networks. The results of the experiments demonstrate that I2B-HGNN excels in diagnosing NDDs with high accuracy, providing interpretable biomarker identification and effective analysis of non-imaging data.
STARFormer: A Novel Spatio-Temporal Aggregation Reorganization Transformer of FMRI for Brain Disorder Diagnosis
Dec 31, 2024



Abstract:Many existing methods that use functional magnetic resonance imaging (fMRI) classify brain disorders, such as autism spectrum disorder (ASD) and attention deficit hyperactivity disorder (ADHD), often overlook the integration of spatial and temporal dependencies of the blood oxygen level-dependent (BOLD) signals, which may lead to inaccurate or imprecise classification results. To solve this problem, we propose a Spatio-Temporal Aggregation eorganization ransformer (STARFormer) that effectively captures both spatial and temporal features of BOLD signals by incorporating three key modules. The region of interest (ROI) spatial structure analysis module uses eigenvector centrality (EC) to reorganize brain regions based on effective connectivity, highlighting critical spatial relationships relevant to the brain disorder. The temporal feature reorganization module systematically segments the time series into equal-dimensional window tokens and captures multiscale features through variable window and cross-window attention. The spatio-temporal feature fusion module employs a parallel transformer architecture with dedicated temporal and spatial branches to extract integrated features. The proposed STARFormer has been rigorously evaluated on two publicly available datasets for the classification of ASD and ADHD. The experimental results confirm that the STARFormer achieves state-of-the-art performance across multiple evaluation metrics, providing a more accurate and reliable tool for the diagnosis of brain disorders and biomedical research. The codes will be available at: https://github.com/NZWANG/STARFormer.
Neural-MCRL: Neural Multimodal Contrastive Representation Learning for EEG-based Visual Decoding
Dec 23, 2024



Abstract:Decoding neural visual representations from electroencephalogram (EEG)-based brain activity is crucial for advancing brain-machine interfaces (BMI) and has transformative potential for neural sensory rehabilitation. While multimodal contrastive representation learning (MCRL) has shown promise in neural decoding, existing methods often overlook semantic consistency and completeness within modalities and lack effective semantic alignment across modalities. This limits their ability to capture the complex representations of visual neural responses. We propose Neural-MCRL, a novel framework that achieves multimodal alignment through semantic bridging and cross-attention mechanisms, while ensuring completeness within modalities and consistency across modalities. Our framework also features the Neural Encoder with Spectral-Temporal Adaptation (NESTA), a EEG encoder that adaptively captures spectral patterns and learns subject-specific transformations. Experimental results demonstrate significant improvements in visual decoding accuracy and model generalization compared to state-of-the-art methods, advancing the field of EEG-based neural visual representation decoding in BMI. Codes will be available at: https://github.com/NZWANG/Neural-MCRL.
Advances in Photoacoustic Imaging Reconstruction and Quantitative Analysis for Biomedical Applications
Nov 05, 2024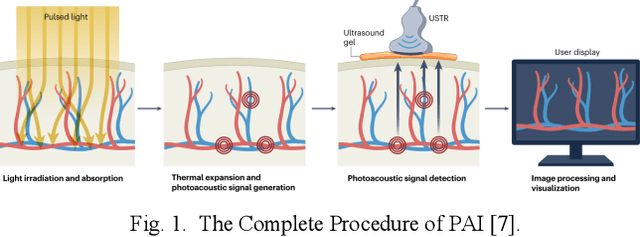
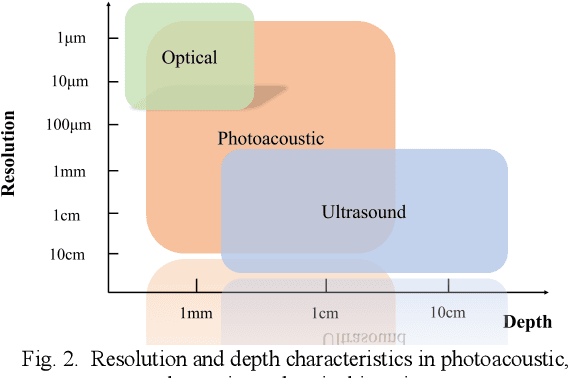
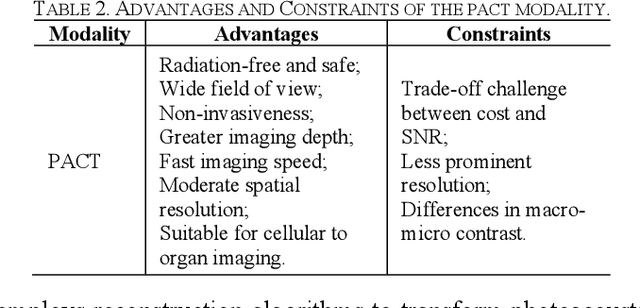
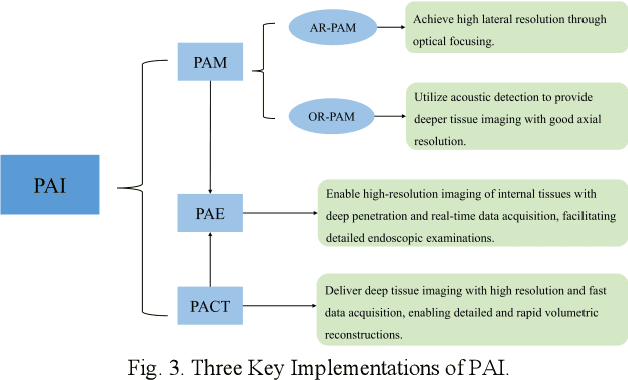
Abstract:Photoacoustic imaging (PAI) represents an innovative biomedical imaging modality that harnesses the advantages of optical resolution and acoustic penetration depth while ensuring enhanced safety. Despite its promising potential across a diverse array of preclinical and clinical applications, the clinical implementation of PAI faces significant challenges, including the trade-off between penetration depth and spatial resolution, as well as the demand for faster imaging speeds. This paper explores the fundamental principles underlying PAI, with a particular emphasis on three primary implementations: photoacoustic computed tomography (PACT), photoacoustic microscopy (PAM), and photoacoustic endoscopy (PAE). We undertake a critical assessment of their respective strengths and practical limitations. Furthermore, recent developments in utilizing conventional or deep learning (DL) methodologies for image reconstruction and artefact mitigation across PACT, PAM, and PAE are outlined, demonstrating considerable potential to enhance image quality and accelerate imaging processes. Furthermore, this paper examines the recent developments in quantitative analysis within PAI, including the quantification of haemoglobin concentration, oxygen saturation, and other physiological parameters within tissues. Finally, our discussion encompasses current trends and future directions in PAI research while emphasizing the transformative impact of deep learning on advancing PAI.
STANet: A Novel Spatio-Temporal Aggregation Network for Depression Classification with Small and Unbalanced FMRI Data
Jul 31, 2024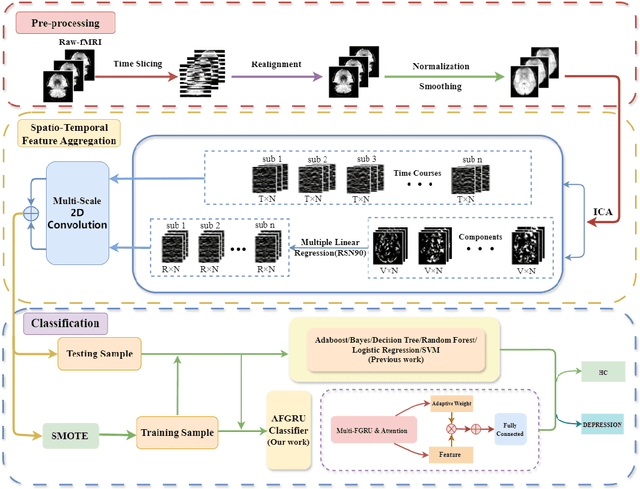
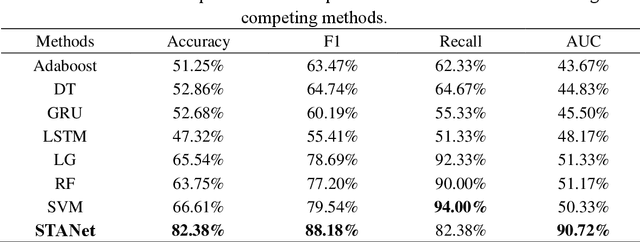
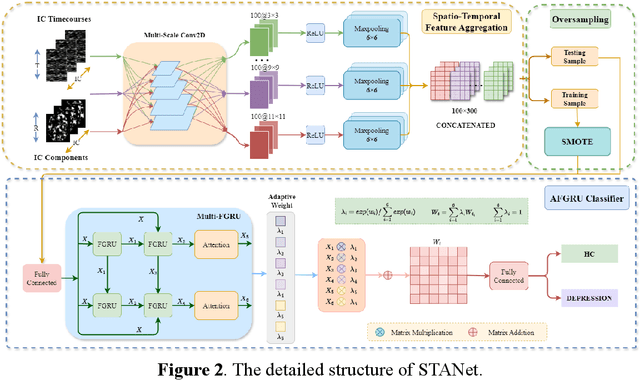
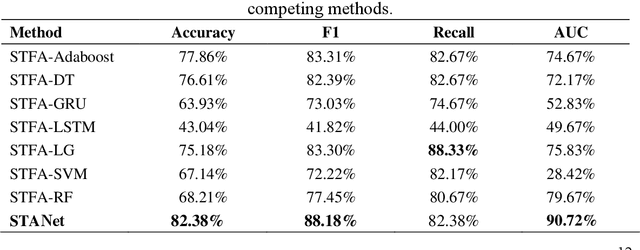
Abstract:Accurate diagnosis of depression is crucial for timely implementation of optimal treatments, preventing complications and reducing the risk of suicide. Traditional methods rely on self-report questionnaires and clinical assessment, lacking objective biomarkers. Combining fMRI with artificial intelligence can enhance depression diagnosis by integrating neuroimaging indicators. However, the specificity of fMRI acquisition for depression often results in unbalanced and small datasets, challenging the sensitivity and accuracy of classification models. In this study, we propose the Spatio-Temporal Aggregation Network (STANet) for diagnosing depression by integrating CNN and RNN to capture both temporal and spatial features of brain activity. STANet comprises the following steps:(1) Aggregate spatio-temporal information via ICA. (2) Utilize multi-scale deep convolution to capture detailed features. (3) Balance data using the SMOTE to generate new samples for minority classes. (4) Employ the AFGRU classifier, which combines Fourier transformation with GRU, to capture long-term dependencies, with an adaptive weight assignment mechanism to enhance model generalization. The experimental results demonstrate that STANet achieves superior depression diagnostic performance with 82.38% accuracy and a 90.72% AUC. The STFA module enhances classification by capturing deeper features at multiple scales. The AFGRU classifier, with adaptive weights and stacked GRU, attains higher accuracy and AUC. SMOTE outperforms other oversampling methods. Additionally, spatio-temporal aggregated features achieve better performance compared to using only temporal or spatial features. STANet outperforms traditional or deep learning classifiers, and functional connectivity-based classifiers, as demonstrated by ten-fold cross-validation.
Neural Modulation Alteration to Positive and Negative Emotions in Depressed Patients: Insights from fMRI Using Positive/Negative Emotion Atlas
Jul 26, 2024
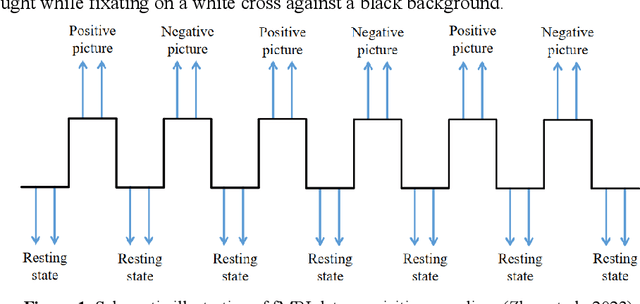


Abstract:Background: Although it has been noticed that depressed patients show differences in processing emotions, the precise neural modulation mechanisms of positive and negative emotions remain elusive. FMRI is a cutting-edge medical imaging technology renowned for its high spatial resolution and dynamic temporal information, making it particularly suitable for the neural dynamics of depression research. Methods: To address this gap, our study firstly leveraged fMRI to delineate activated regions associated with positive and negative emotions in healthy individuals, resulting in the creation of positive emotion atlas (PEA) and negative emotion atlas (NEA). Subsequently, we examined neuroimaging changes in depression patients using these atlases and evaluated their diagnostic performance based on machine learning. Results: Our findings demonstrate that the classification accuracy of depressed patients based on PEA and NEA exceeded 0.70, a notable improvement compared to the whole-brain atlases. Furthermore, ALFF analysis unveiled significant differences between depressed patients and healthy controls in eight functional clusters during the NEA, focusing on the left cuneus, cingulate gyrus, and superior parietal lobule. In contrast, the PEA revealed more pronounced differences across fifteen clusters, involving the right fusiform gyrus, parahippocampal gyrus, and inferior parietal lobule. Limitations: Due to the limited sample size and subtypes of depressed patients, the efficacy may need further validation in future. Conclusions: These findings emphasize the complex interplay between emotion modulation and depression, showcasing significant alterations in both PEA and NEA among depression patients. This research enhances our understanding of emotion modulation in depression, with implications for diagnosis and treatment evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge