Luhui Cai
MHNet: Multi-view High-order Network for Diagnosing Neurodevelopmental Disorders Using Resting-state fMRI
Jul 03, 2024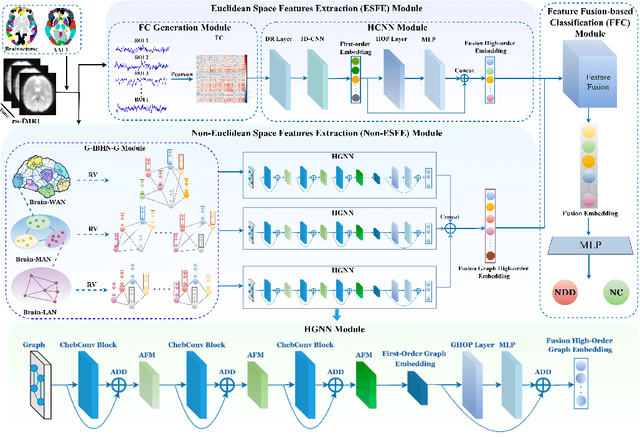
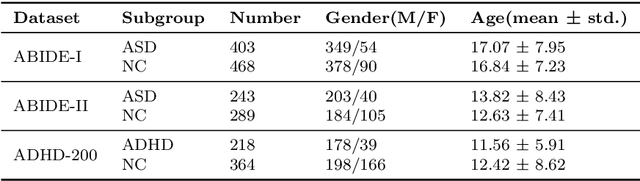
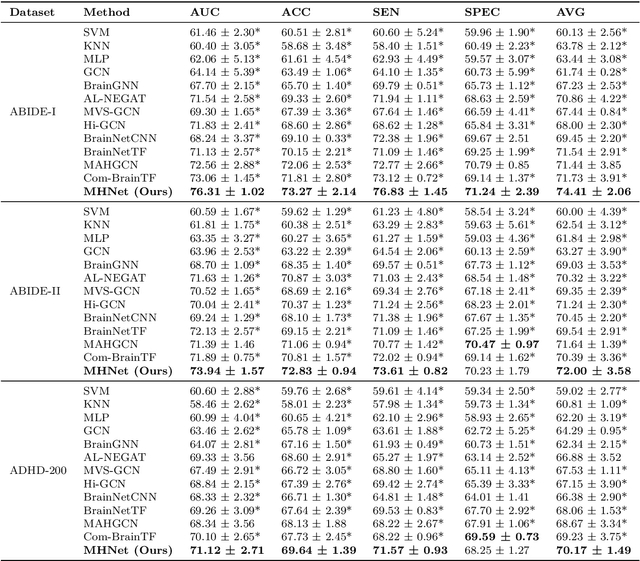
Abstract:Background: Deep learning models have shown promise in diagnosing neurodevelopmental disorders (NDD) like ASD and ADHD. However, many models either use graph neural networks (GNN) to construct single-level brain functional networks (BFNs) or employ spatial convolution filtering for local information extraction from rs-fMRI data, often neglecting high-order features crucial for NDD classification. Methods: We introduce a Multi-view High-order Network (MHNet) to capture hierarchical and high-order features from multi-view BFNs derived from rs-fMRI data for NDD prediction. MHNet has two branches: the Euclidean Space Features Extraction (ESFE) module and the Non-Euclidean Space Features Extraction (Non-ESFE) module, followed by a Feature Fusion-based Classification (FFC) module for NDD identification. ESFE includes a Functional Connectivity Generation (FCG) module and a High-order Convolutional Neural Network (HCNN) module to extract local and high-order features from BFNs in Euclidean space. Non-ESFE comprises a Generic Internet-like Brain Hierarchical Network Generation (G-IBHN-G) module and a High-order Graph Neural Network (HGNN) module to capture topological and high-order features in non-Euclidean space. Results: Experiments on three public datasets show that MHNet outperforms state-of-the-art methods using both AAL1 and Brainnetome Atlas templates. Extensive ablation studies confirm the superiority of MHNet and the effectiveness of using multi-view fMRI information and high-order features. Our study also offers atlas options for constructing more sophisticated hierarchical networks and explains the association between key brain regions and NDD. Conclusion: MHNet leverages multi-view feature learning from both Euclidean and non-Euclidean spaces, incorporating high-order information from BFNs to enhance NDD classification performance.
You Only Acquire Sparse-channel (YOAS): A Unified Framework for Dense-channel EEG Generation
Jun 21, 2024



Abstract:High-precision acquisition of dense-channel electroencephalogram (EEG) signals is often impeded by the costliness and lack of portability of equipment. In contrast, generating dense-channel EEG signals effectively from sparse channels shows promise and economic viability. However, sparse-channel EEG poses challenges such as reduced spatial resolution, information loss, signal mixing, and heightened susceptibility to noise and interference. To address these challenges, we first theoretically formulate the dense-channel EEG generation problem as by optimizing a set of cross-channel EEG signal generation problems. Then, we propose the YOAS framework for generating dense-channel data from sparse-channel EEG signals. The YOAS totally consists of four sequential stages: Data Preparation, Data Preprocessing, Biased-EEG Generation, and Synthetic EEG Generation. Data Preparation and Preprocessing carefully consider the distribution of EEG electrodes and low signal-to-noise ratio problem of EEG signals. Biased-EEG Generation includes sub-modules of BiasEEGGanFormer and BiasEEGDiffFormer, which facilitate long-term feature extraction with attention and generate signals by combining electrode position alignment with diffusion model, respectively. Synthetic EEG Generation synthesizes the final signals, employing a deduction paradigm for multi-channel EEG generation. Extensive experiments confirmed YOAS's feasibility, efficiency, and theoretical validity, even remarkably enhancing data discernibility. This breakthrough in dense-channel EEG signal generation from sparse-channel data opens new avenues for exploration in EEG signal processing and application.
MM-GTUNets: Unified Multi-Modal Graph Deep Learning for Brain Disorders Prediction
Jun 20, 2024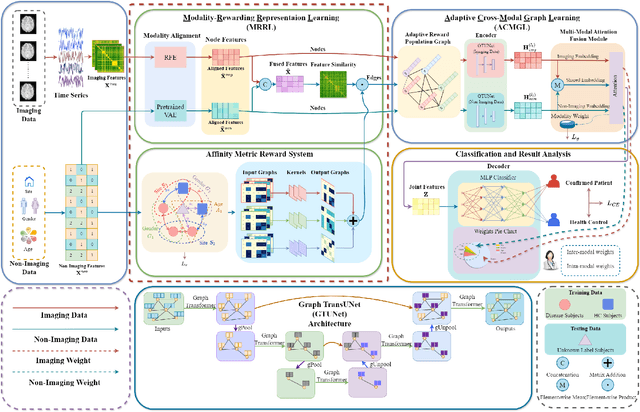

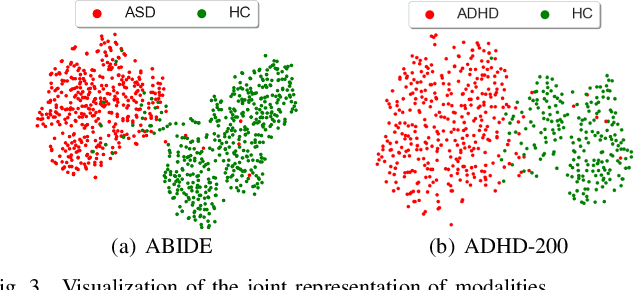
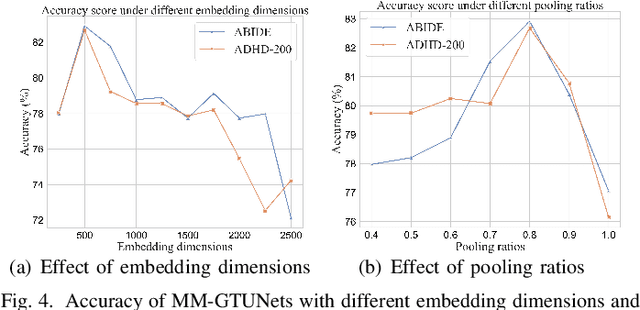
Abstract:Graph deep learning (GDL) has demonstrated impressive performance in predicting population-based brain disorders (BDs) through the integration of both imaging and non-imaging data. However, the effectiveness of GDL based methods heavily depends on the quality of modeling the multi-modal population graphs and tends to degrade as the graph scale increases. Furthermore, these methods often constrain interactions between imaging and non-imaging data to node-edge interactions within the graph, overlooking complex inter-modal correlations, leading to suboptimal outcomes. To overcome these challenges, we propose MM-GTUNets, an end-to-end graph transformer based multi-modal graph deep learning (MMGDL) framework designed for brain disorders prediction at large scale. Specifically, to effectively leverage rich multi-modal information related to diseases, we introduce Modality Reward Representation Learning (MRRL) which adaptively constructs population graphs using a reward system. Additionally, we employ variational autoencoder to reconstruct latent representations of non-imaging features aligned with imaging features. Based on this, we propose Adaptive Cross-Modal Graph Learning (ACMGL), which captures critical modality-specific and modality-shared features through a unified GTUNet encoder taking advantages of Graph UNet and Graph Transformer, and feature fusion module. We validated our method on two public multi-modal datasets ABIDE and ADHD-200, demonstrating its superior performance in diagnosing BDs. Our code is available at https://github.com/NZWANG/MM-GTUNets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge