Lingbin Bian
Information Bottleneck-Guided Heterogeneous Graph Learning for Interpretable Neurodevelopmental Disorder Diagnosis
Feb 28, 2025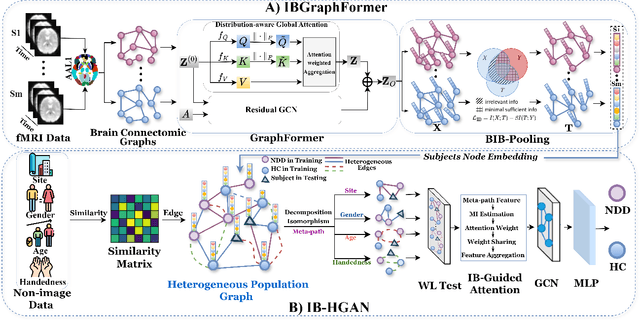
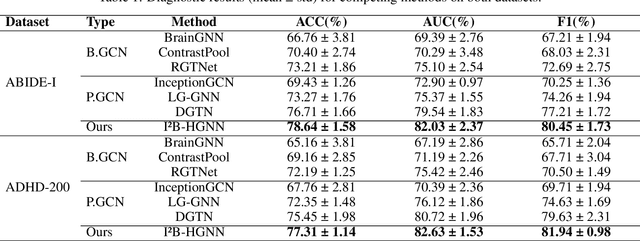
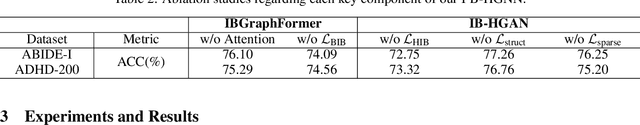
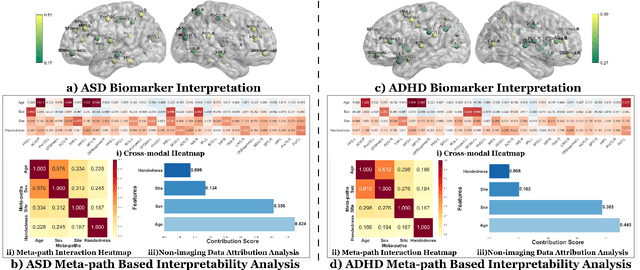
Abstract:Developing interpretable models for diagnosing neurodevelopmental disorders (NDDs) is highly valuable yet challenging, primarily due to the complexity of encoding, decoding and integrating imaging and non-imaging data. Many existing machine learning models struggle to provide comprehensive interpretability, often failing to extract meaningful biomarkers from imaging data, such as functional magnetic resonance imaging (fMRI), or lacking mechanisms to explain the significance of non-imaging data. In this paper, we propose the Interpretable Information Bottleneck Heterogeneous Graph Neural Network (I2B-HGNN), a novel framework designed to learn from fine-grained local patterns to comprehensive global multi-modal interactions. This framework comprises two key modules. The first module, the Information Bottleneck Graph Transformer (IBGraphFormer) for local patterns, integrates global modeling with brain connectomic-constrained graph neural networks to identify biomarkers through information bottleneck-guided pooling. The second module, the Information Bottleneck Heterogeneous Graph Attention Network (IB-HGAN) for global multi-modal interactions, facilitates interpretable multi-modal fusion of imaging and non-imaging data using heterogeneous graph neural networks. The results of the experiments demonstrate that I2B-HGNN excels in diagnosing NDDs with high accuracy, providing interpretable biomarker identification and effective analysis of non-imaging data.
MHNet: Multi-view High-order Network for Diagnosing Neurodevelopmental Disorders Using Resting-state fMRI
Jul 03, 2024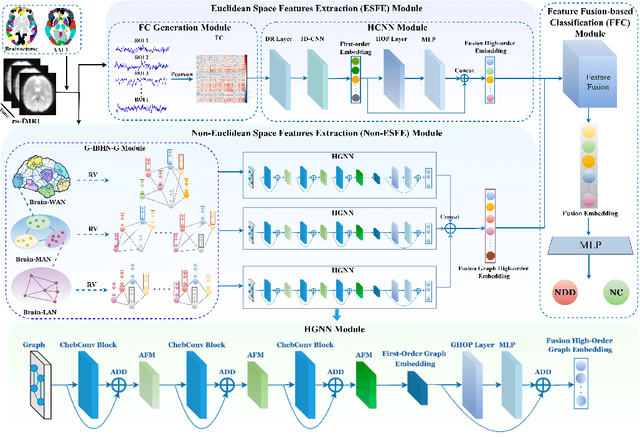
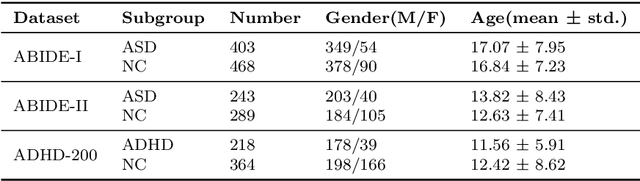
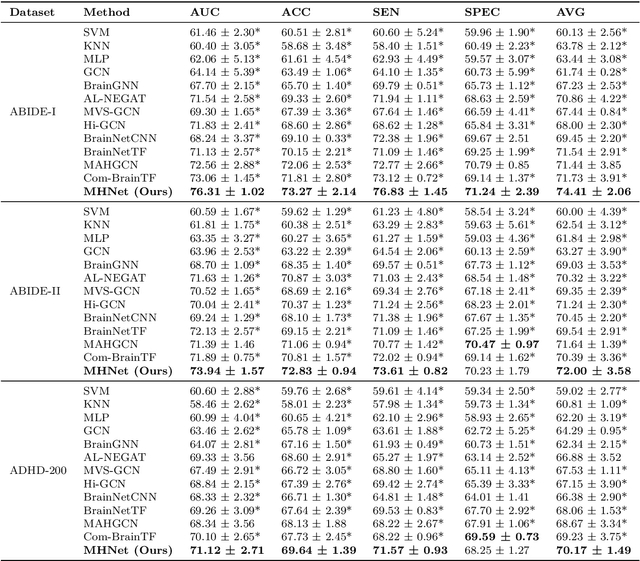
Abstract:Background: Deep learning models have shown promise in diagnosing neurodevelopmental disorders (NDD) like ASD and ADHD. However, many models either use graph neural networks (GNN) to construct single-level brain functional networks (BFNs) or employ spatial convolution filtering for local information extraction from rs-fMRI data, often neglecting high-order features crucial for NDD classification. Methods: We introduce a Multi-view High-order Network (MHNet) to capture hierarchical and high-order features from multi-view BFNs derived from rs-fMRI data for NDD prediction. MHNet has two branches: the Euclidean Space Features Extraction (ESFE) module and the Non-Euclidean Space Features Extraction (Non-ESFE) module, followed by a Feature Fusion-based Classification (FFC) module for NDD identification. ESFE includes a Functional Connectivity Generation (FCG) module and a High-order Convolutional Neural Network (HCNN) module to extract local and high-order features from BFNs in Euclidean space. Non-ESFE comprises a Generic Internet-like Brain Hierarchical Network Generation (G-IBHN-G) module and a High-order Graph Neural Network (HGNN) module to capture topological and high-order features in non-Euclidean space. Results: Experiments on three public datasets show that MHNet outperforms state-of-the-art methods using both AAL1 and Brainnetome Atlas templates. Extensive ablation studies confirm the superiority of MHNet and the effectiveness of using multi-view fMRI information and high-order features. Our study also offers atlas options for constructing more sophisticated hierarchical networks and explains the association between key brain regions and NDD. Conclusion: MHNet leverages multi-view feature learning from both Euclidean and non-Euclidean spaces, incorporating high-order information from BFNs to enhance NDD classification performance.
MM-GTUNets: Unified Multi-Modal Graph Deep Learning for Brain Disorders Prediction
Jun 20, 2024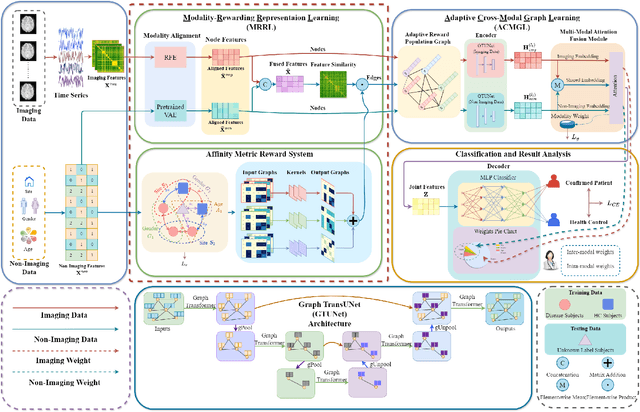

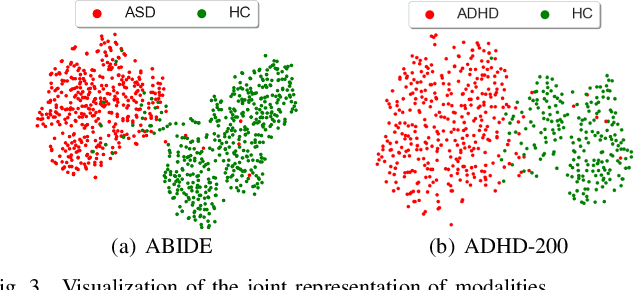
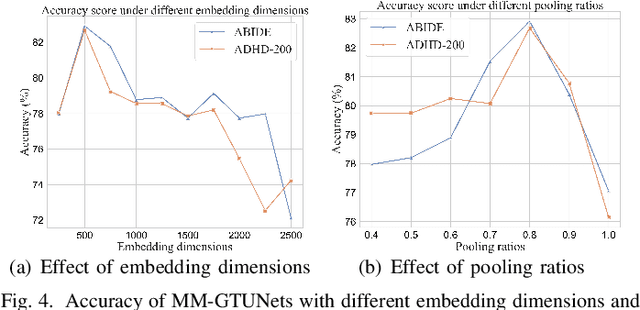
Abstract:Graph deep learning (GDL) has demonstrated impressive performance in predicting population-based brain disorders (BDs) through the integration of both imaging and non-imaging data. However, the effectiveness of GDL based methods heavily depends on the quality of modeling the multi-modal population graphs and tends to degrade as the graph scale increases. Furthermore, these methods often constrain interactions between imaging and non-imaging data to node-edge interactions within the graph, overlooking complex inter-modal correlations, leading to suboptimal outcomes. To overcome these challenges, we propose MM-GTUNets, an end-to-end graph transformer based multi-modal graph deep learning (MMGDL) framework designed for brain disorders prediction at large scale. Specifically, to effectively leverage rich multi-modal information related to diseases, we introduce Modality Reward Representation Learning (MRRL) which adaptively constructs population graphs using a reward system. Additionally, we employ variational autoencoder to reconstruct latent representations of non-imaging features aligned with imaging features. Based on this, we propose Adaptive Cross-Modal Graph Learning (ACMGL), which captures critical modality-specific and modality-shared features through a unified GTUNet encoder taking advantages of Graph UNet and Graph Transformer, and feature fusion module. We validated our method on two public multi-modal datasets ABIDE and ADHD-200, demonstrating its superior performance in diagnosing BDs. Our code is available at https://github.com/NZWANG/MM-GTUNets.
WSEBP: A Novel Width-depth Synchronous Extension-based Basis Pursuit Algorithm for Multi-Layer Convolutional Sparse Coding
Mar 30, 2022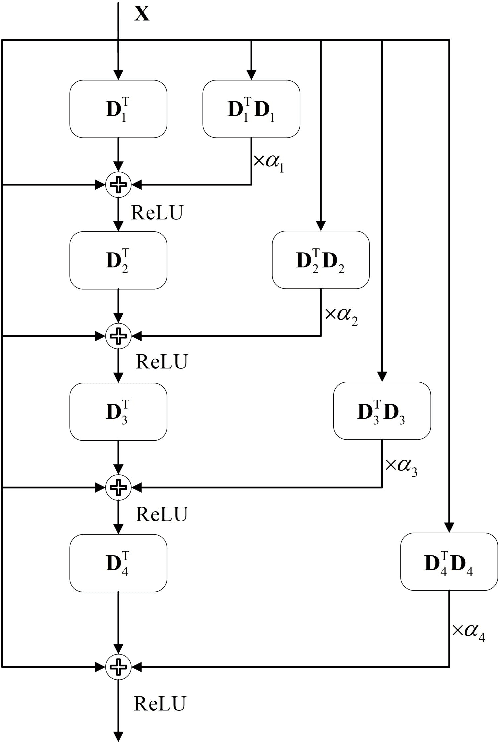
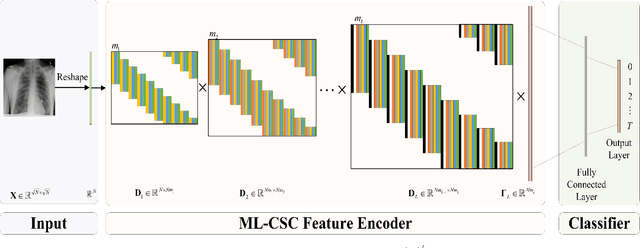
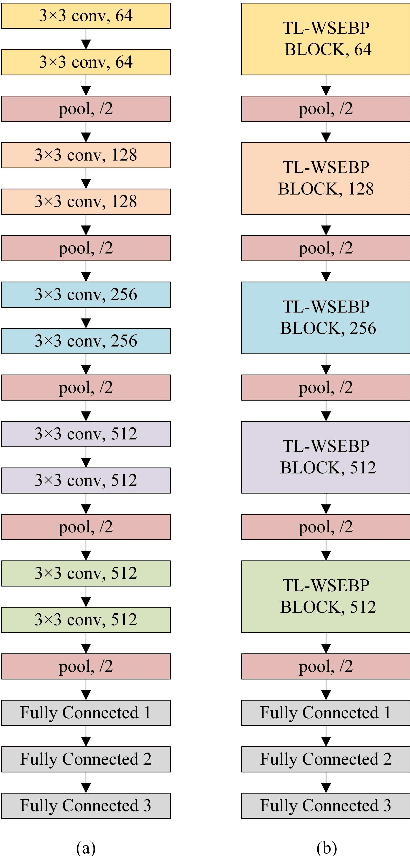
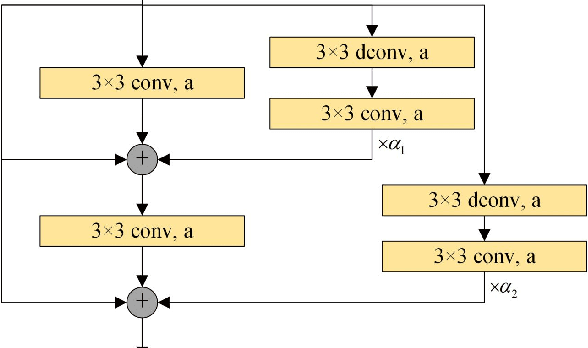
Abstract:The pursuit algorithms integrated in multi-layer convolutional sparse coding (ML-CSC) can interpret the convolutional neural networks (CNNs). However, many current state-of-art (SOTA) pursuit algorithms require multiple iterations to optimize the solution of ML-CSC, which limits their applications to deeper CNNs due to high computational cost and large number of resources for getting very tiny gain of performance. In this study, we focus on the 0th iteration in pursuit algorithm by introducing an effective initialization strategy for each layer, by which the solution for ML-CSC can be improved. Specifically, we first propose a novel width-depth synchronous extension-based basis pursuit (WSEBP) algorithm which solves the ML-CSC problem without the limitation of the number of iterations compared to the SOTA algorithms and maximizes the performance by an effective initialization in each layer. Then, we propose a simple and unified ML-CSC-based classification network (ML-CSC-Net) which consists of an ML-CSC-based feature encoder and a fully-connected layer to validate the performance of WSEBP on image classification task. The experimental results show that our proposed WSEBP outperforms SOTA algorithms in terms of accuracy and consumption resources. In addition, the WSEBP integrated in CNNs can improve the performance of deeper CNNs and make them interpretable. Finally, taking VGG as an example, we propose WSEBP-VGG13 to enhance the performance of VGG13, which achieves competitive results on four public datasets, i.e., 87.79% vs. 86.83% on Cifar-10 dataset, 58.01% vs. 54.60% on Cifar-100 dataset, 91.52% vs. 89.58% on COVID-19 dataset, and 99.88% vs. 99.78% on Crack dataset, respectively. The results show the effectiveness of the proposed WSEBP, the improved performance of ML-CSC with WSEBP, and interpretation of the CNNs or deeper CNNs.
Identification of brain states, transitions, and communities using functional MRI
Jan 26, 2021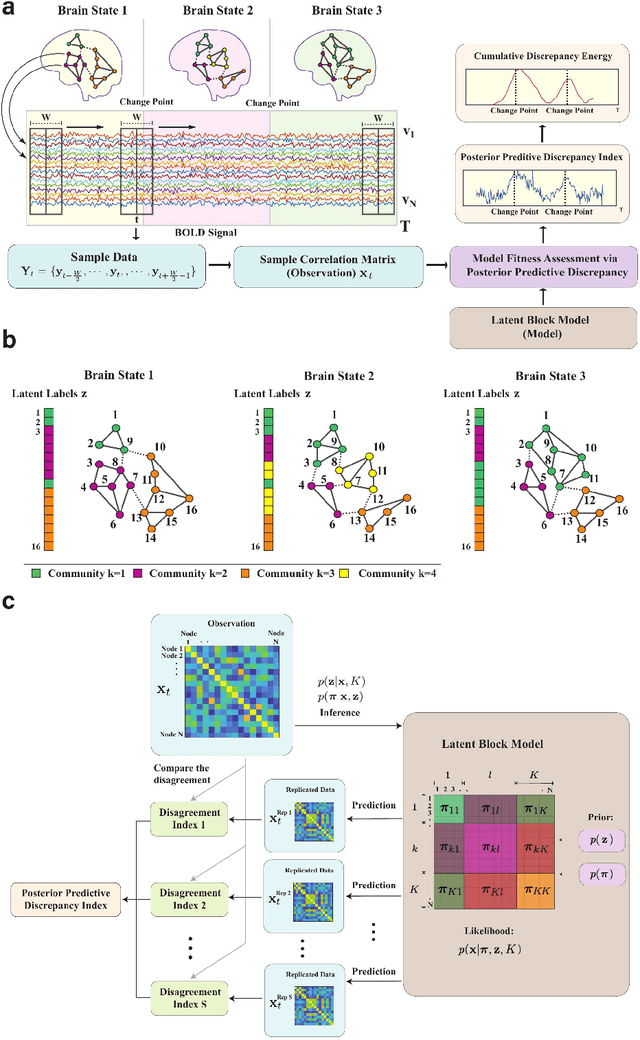
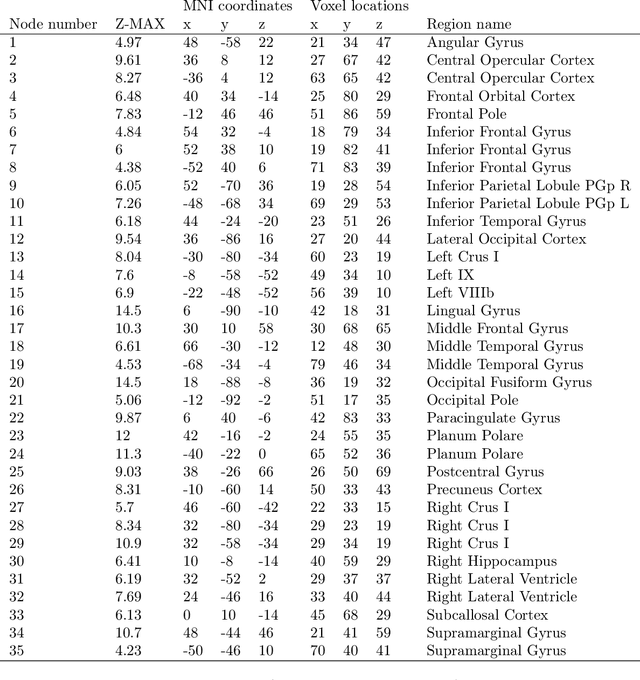
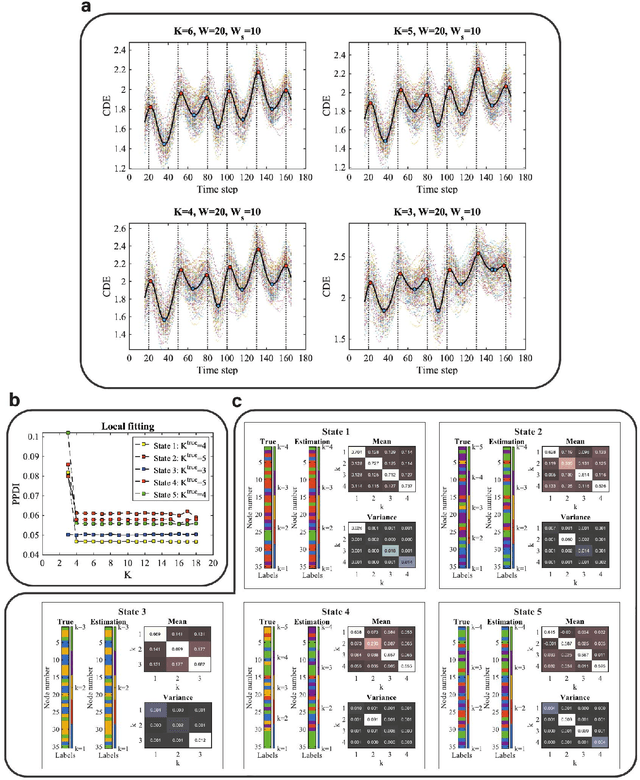
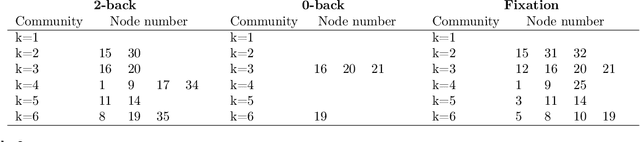
Abstract:Brain function relies on a precisely coordinated and dynamic balance between the functional integration and segregation of distinct neural systems. Characterizing the way in which neural systems reconfigure their interactions to give rise to distinct but hidden brain states remains an open challenge. In this paper, we propose a Bayesian model-based characterization of latent brain states and showcase a novel method based on posterior predictive discrepancy using the latent block model to detect transitions between latent brain states in blood oxygen level-dependent (BOLD) time series. The set of estimated parameters in the model includes a latent label vector that assigns network nodes to communities, and also block model parameters that reflect the weighted connectivity within and between communities. Besides extensive in-silico model evaluation, we also provide empirical validation (and replication) using the Human Connectome Project (HCP) dataset of 100 healthy adults. Our results obtained through an analysis of task-fMRI data during working memory performance show appropriate lags between external task demands and change-points between brain states, with distinctive community patterns distinguishing fixation, low-demand and high-demand task conditions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge