Kaile Zhang
STANet: A Novel Spatio-Temporal Aggregation Network for Depression Classification with Small and Unbalanced FMRI Data
Jul 31, 2024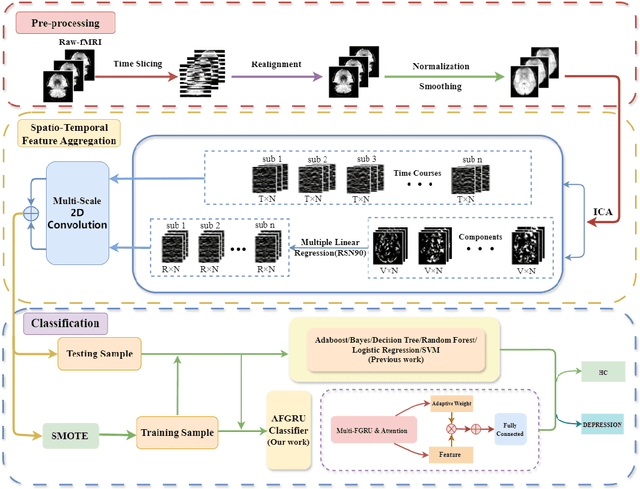
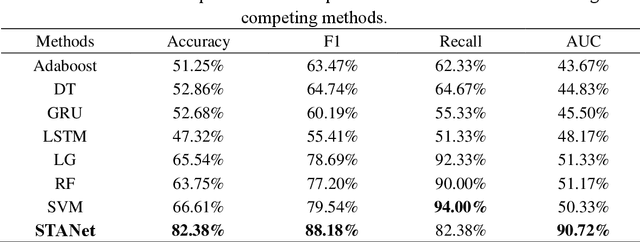
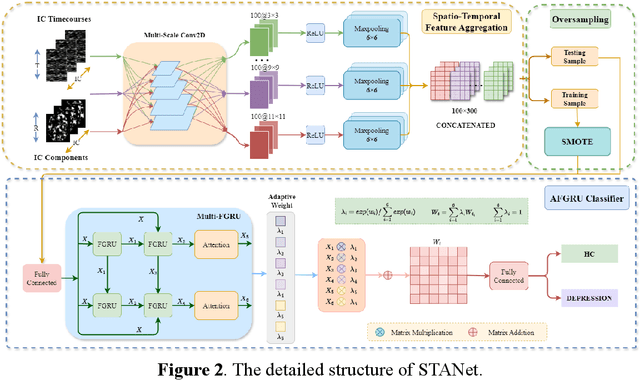
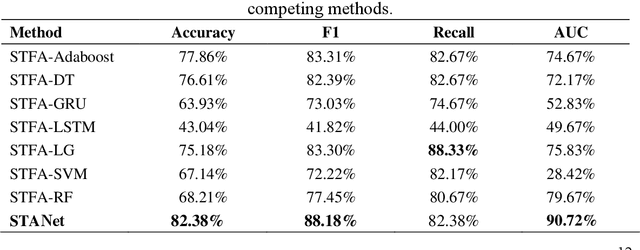
Abstract:Accurate diagnosis of depression is crucial for timely implementation of optimal treatments, preventing complications and reducing the risk of suicide. Traditional methods rely on self-report questionnaires and clinical assessment, lacking objective biomarkers. Combining fMRI with artificial intelligence can enhance depression diagnosis by integrating neuroimaging indicators. However, the specificity of fMRI acquisition for depression often results in unbalanced and small datasets, challenging the sensitivity and accuracy of classification models. In this study, we propose the Spatio-Temporal Aggregation Network (STANet) for diagnosing depression by integrating CNN and RNN to capture both temporal and spatial features of brain activity. STANet comprises the following steps:(1) Aggregate spatio-temporal information via ICA. (2) Utilize multi-scale deep convolution to capture detailed features. (3) Balance data using the SMOTE to generate new samples for minority classes. (4) Employ the AFGRU classifier, which combines Fourier transformation with GRU, to capture long-term dependencies, with an adaptive weight assignment mechanism to enhance model generalization. The experimental results demonstrate that STANet achieves superior depression diagnostic performance with 82.38% accuracy and a 90.72% AUC. The STFA module enhances classification by capturing deeper features at multiple scales. The AFGRU classifier, with adaptive weights and stacked GRU, attains higher accuracy and AUC. SMOTE outperforms other oversampling methods. Additionally, spatio-temporal aggregated features achieve better performance compared to using only temporal or spatial features. STANet outperforms traditional or deep learning classifiers, and functional connectivity-based classifiers, as demonstrated by ten-fold cross-validation.
Neural Modulation Alteration to Positive and Negative Emotions in Depressed Patients: Insights from fMRI Using Positive/Negative Emotion Atlas
Jul 26, 2024
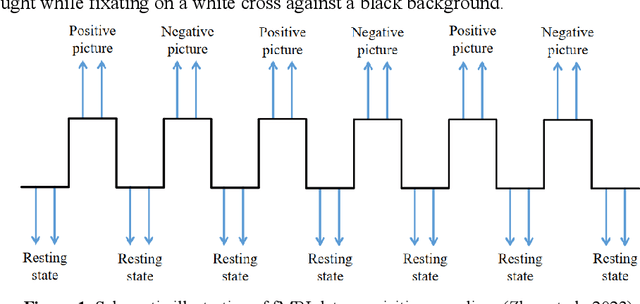


Abstract:Background: Although it has been noticed that depressed patients show differences in processing emotions, the precise neural modulation mechanisms of positive and negative emotions remain elusive. FMRI is a cutting-edge medical imaging technology renowned for its high spatial resolution and dynamic temporal information, making it particularly suitable for the neural dynamics of depression research. Methods: To address this gap, our study firstly leveraged fMRI to delineate activated regions associated with positive and negative emotions in healthy individuals, resulting in the creation of positive emotion atlas (PEA) and negative emotion atlas (NEA). Subsequently, we examined neuroimaging changes in depression patients using these atlases and evaluated their diagnostic performance based on machine learning. Results: Our findings demonstrate that the classification accuracy of depressed patients based on PEA and NEA exceeded 0.70, a notable improvement compared to the whole-brain atlases. Furthermore, ALFF analysis unveiled significant differences between depressed patients and healthy controls in eight functional clusters during the NEA, focusing on the left cuneus, cingulate gyrus, and superior parietal lobule. In contrast, the PEA revealed more pronounced differences across fifteen clusters, involving the right fusiform gyrus, parahippocampal gyrus, and inferior parietal lobule. Limitations: Due to the limited sample size and subtypes of depressed patients, the efficacy may need further validation in future. Conclusions: These findings emphasize the complex interplay between emotion modulation and depression, showcasing significant alterations in both PEA and NEA among depression patients. This research enhances our understanding of emotion modulation in depression, with implications for diagnosis and treatment evaluation.
Puzzle Pieces Picker: Deciphering Ancient Chinese Characters with Radical Reconstruction
Jun 05, 2024



Abstract:Oracle Bone Inscriptions is one of the oldest existing forms of writing in the world. However, due to the great antiquity of the era, a large number of Oracle Bone Inscriptions (OBI) remain undeciphered, making it one of the global challenges in the field of paleography today. This paper introduces a novel approach, namely Puzzle Pieces Picker (P$^3$), to decipher these enigmatic characters through radical reconstruction. We deconstruct OBI into foundational strokes and radicals, then employ a Transformer model to reconstruct them into their modern (conterpart)\textcolor{blue}{counterparts}, offering a groundbreaking solution to ancient script analysis. To further this endeavor, a new Ancient Chinese Character Puzzles (ACCP) dataset was developed, comprising an extensive collection of character images from seven key historical stages, annotated with detailed radical sequences. The experiments have showcased considerable promising insights, underscoring the potential and effectiveness of our approach in deciphering the intricacies of ancient Chinese scripts. Through this novel dataset and methodology, we aim to bridge the gap between traditional philology and modern document analysis techniques, offering new insights into the rich history of Chinese linguistic heritage.
An open dataset for oracle bone script recognition and decipherment
Jan 27, 2024Abstract:Oracle Bone Script (OBS), one of the earliest known forms of ancient Chinese writing, holds invaluable insights into the humanities and geography of the Shang Dynasty, dating back 3,000 years. The immense historical and cultural significance of these writings cannot be overstated. However, the passage of time has obscured much of their meaning, presenting a significant challenge in deciphering these ancient texts. With the advent of Artificial Intelligence (AI), employing AI to assist in interpreting OBS has become a feasible option. Yet, progress in this area has been hindered by a lack of high-quality datasets. To address this issue, this paper details the creation of the HUST-OBS dataset. This dataset encompasses 77,064 images of 1,588 individual deciphered scripts and 62,989 images of 9,411 undeciphered characters, with a total of 140,053 images, compiled from diverse sources. Additionally, all images and labels have been reviewed and corrected by experts in oracle bone studies. The hope is that this dataset could inspire and assist future research in deciphering those unknown OBS.
An open dataset for the evolution of oracle bone characters: EVOBC
Jan 23, 2024Abstract:The earliest extant Chinese characters originate from oracle bone inscriptions, which are closely related to other East Asian languages. These inscriptions hold immense value for anthropology and archaeology. However, deciphering oracle bone script remains a formidable challenge, with only approximately 1,600 of the over 4,500 extant characters elucidated to date. Further scholarly investigation is required to comprehensively understand this ancient writing system. Artificial Intelligence technology is a promising avenue for deciphering oracle bone characters, particularly concerning their evolution. However, one of the challenges is the lack of datasets mapping the evolution of these characters over time. In this study, we systematically collected ancient characters from authoritative texts and websites spanning six historical stages: Oracle Bone Characters - OBC (15th century B.C.), Bronze Inscriptions - BI (13th to 221 B.C.), Seal Script - SS (11th to 8th centuries B.C.), Spring and Autumn period Characters - SAC (770 to 476 B.C.), Warring States period Characters - WSC (475 B.C. to 221 B.C.), and Clerical Script - CS (221 B.C. to 220 A.D.). Subsequently, we constructed an extensive dataset, namely EVolution Oracle Bone Characters (EVOBC), consisting of 229,170 images representing 13,714 distinct character categories. We conducted validation and simulated deciphering on the constructed dataset, and the results demonstrate its high efficacy in aiding the study of oracle bone script. This openly accessible dataset aims to digitalize ancient Chinese scripts across multiple eras, facilitating the decipherment of oracle bone script by examining the evolution of glyph forms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge