Pedro R. A. S. Bassi
Early and Prediagnostic Detection of Pancreatic Cancer from Computed Tomography
Jan 29, 2026Abstract:Pancreatic ductal adenocarcinoma (PDAC), one of the deadliest solid malignancies, is often detected at a late and inoperable stage. Retrospective reviews of prediagnostic CT scans, when conducted by expert radiologists aware that the patient later developed PDAC, frequently reveal lesions that were previously overlooked. To help detecting these lesions earlier, we developed an automated system named ePAI (early Pancreatic cancer detection with Artificial Intelligence). It was trained on data from 1,598 patients from a single medical center. In the internal test involving 1,009 patients, ePAI achieved an area under the receiver operating characteristic curve (AUC) of 0.939-0.999, a sensitivity of 95.3%, and a specificity of 98.7% for detecting small PDAC less than 2 cm in diameter, precisely localizing PDAC as small as 2 mm. In an external test involving 7,158 patients across 6 centers, ePAI achieved an AUC of 0.918-0.945, a sensitivity of 91.5%, and a specificity of 88.0%, precisely localizing PDAC as small as 5 mm. Importantly, ePAI detected PDACs on prediagnostic CT scans obtained 3 to 36 months before clinical diagnosis that had originally been overlooked by radiologists. It successfully detected and localized PDACs in 75 of 159 patients, with a median lead time of 347 days before clinical diagnosis. Our multi-reader study showed that ePAI significantly outperformed 30 board-certified radiologists by 50.3% (P < 0.05) in sensitivity while maintaining a comparable specificity of 95.4% in detecting PDACs early and prediagnostic. These findings suggest its potential of ePAI as an assistive tool to improve early detection of pancreatic cancer.
Auditing Significance, Metric Choice, and Demographic Fairness in Medical AI Challenges
Dec 22, 2025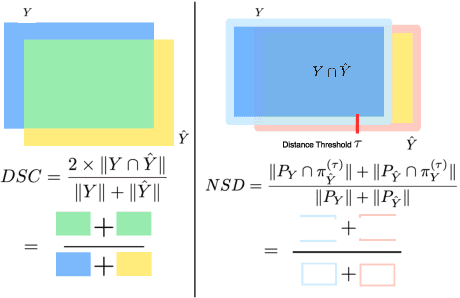
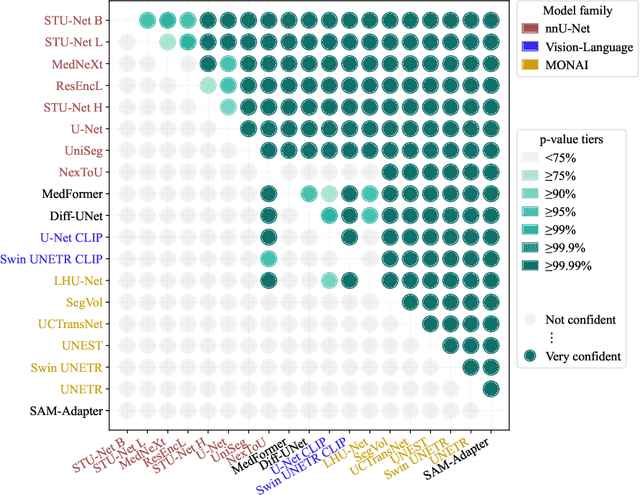
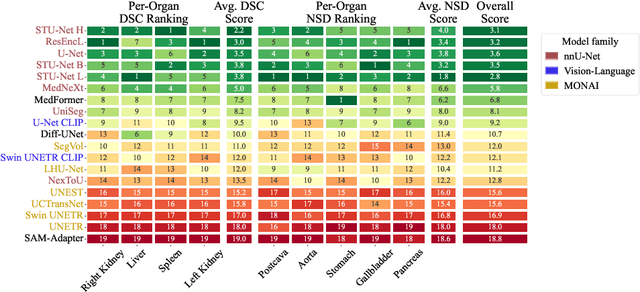
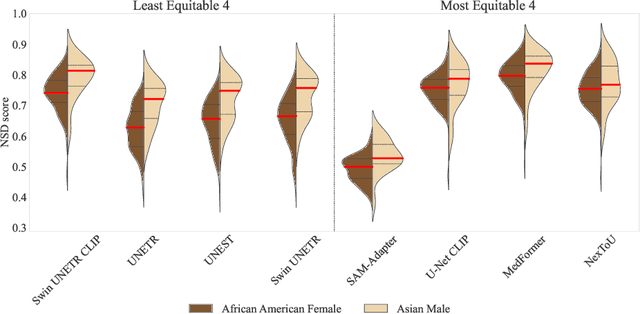
Abstract:Open challenges have become the de facto standard for comparative ranking of medical AI methods. Despite their importance, medical AI leaderboards exhibit three persistent limitations: (1) score gaps are rarely tested for statistical significance, so rank stability is unknown; (2) single averaged metrics are applied to every organ, hiding clinically important boundary errors; (3) performance across intersecting demographics is seldom reported, masking fairness and equity gaps. We introduce RankInsight, an open-source toolkit that seeks to address these limitations. RankInsight (1) computes pair-wise significance maps that show the nnU-Net family outperforms Vision-Language and MONAI submissions with high statistical certainty; (2) recomputes leaderboards with organ-appropriate metrics, reversing the order of the top four models when Dice is replaced by NSD for tubular structures; and (3) audits intersectional fairness, revealing that more than half of the MONAI-based entries have the largest gender-race discrepancy on our proprietary Johns Hopkins Hospital dataset. The RankInsight toolkit is publicly released and can be directly applied to past, ongoing, and future challenges. It enables organizers and participants to publish rankings that are statistically sound, clinically meaningful, and demographically fair.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Abstract:Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
PanTS: The Pancreatic Tumor Segmentation Dataset
Jul 02, 2025Abstract:PanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
Are Vision Language Models Ready for Clinical Diagnosis? A 3D Medical Benchmark for Tumor-centric Visual Question Answering
May 25, 2025Abstract:Vision-Language Models (VLMs) have shown promise in various 2D visual tasks, yet their readiness for 3D clinical diagnosis remains unclear due to stringent demands for recognition precision, reasoning ability, and domain knowledge. To systematically evaluate these dimensions, we present DeepTumorVQA, a diagnostic visual question answering (VQA) benchmark targeting abdominal tumors in CT scans. It comprises 9,262 CT volumes (3.7M slices) from 17 public datasets, with 395K expert-level questions spanning four categories: Recognition, Measurement, Visual Reasoning, and Medical Reasoning. DeepTumorVQA introduces unique challenges, including small tumor detection and clinical reasoning across 3D anatomy. Benchmarking four advanced VLMs (RadFM, M3D, Merlin, CT-CHAT), we find current models perform adequately on measurement tasks but struggle with lesion recognition and reasoning, and are still not meeting clinical needs. Two key insights emerge: (1) large-scale multimodal pretraining plays a crucial role in DeepTumorVQA testing performance, making RadFM stand out among all VLMs. (2) Our dataset exposes critical differences in VLM components, where proper image preprocessing and design of vision modules significantly affect 3D perception. To facilitate medical multimodal research, we have released DeepTumorVQA as a rigorous benchmark: https://github.com/Schuture/DeepTumorVQA.
Text2CT: Towards 3D CT Volume Generation from Free-text Descriptions Using Diffusion Model
May 07, 2025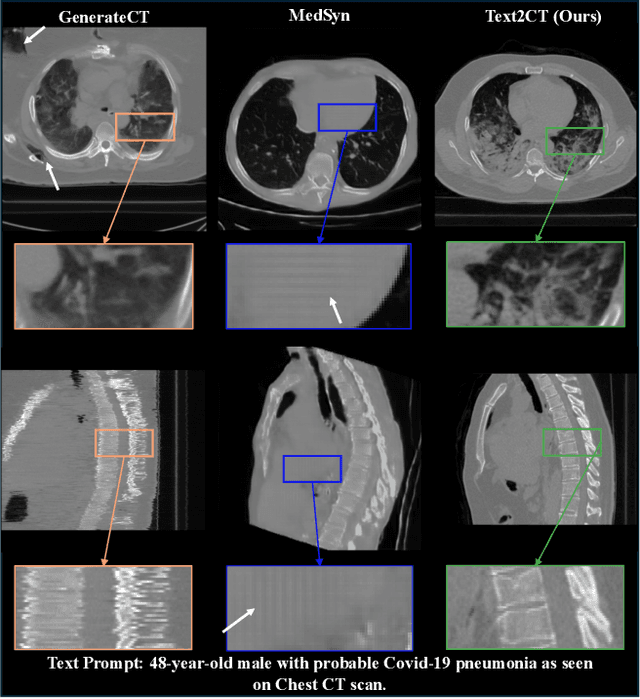
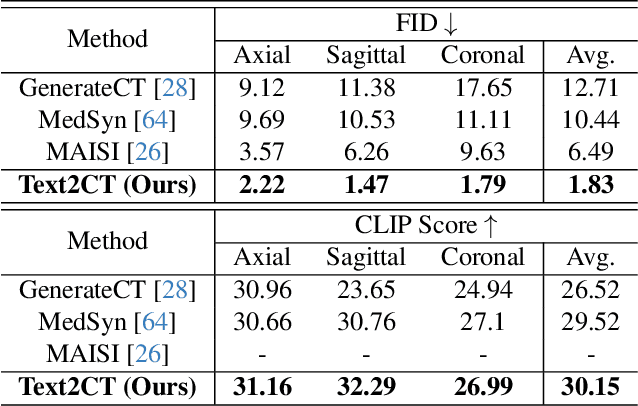
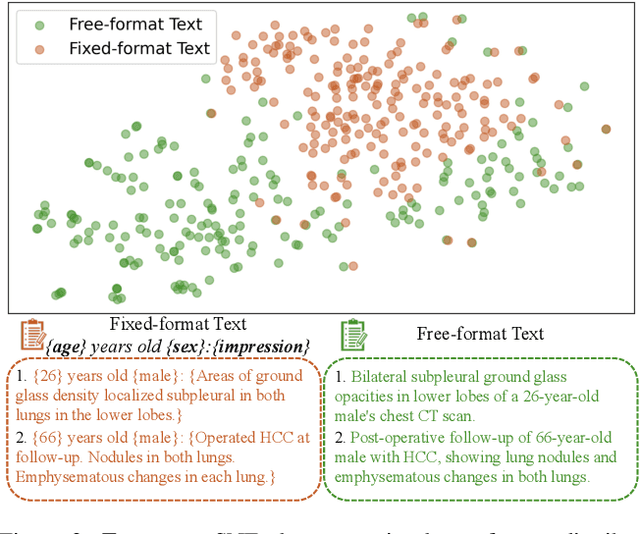
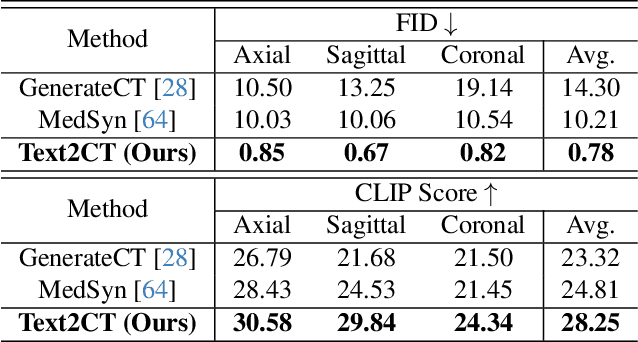
Abstract:Generating 3D CT volumes from descriptive free-text inputs presents a transformative opportunity in diagnostics and research. In this paper, we introduce Text2CT, a novel approach for synthesizing 3D CT volumes from textual descriptions using the diffusion model. Unlike previous methods that rely on fixed-format text input, Text2CT employs a novel prompt formulation that enables generation from diverse, free-text descriptions. The proposed framework encodes medical text into latent representations and decodes them into high-resolution 3D CT scans, effectively bridging the gap between semantic text inputs and detailed volumetric representations in a unified 3D framework. Our method demonstrates superior performance in preserving anatomical fidelity and capturing intricate structures as described in the input text. Extensive evaluations show that our approach achieves state-of-the-art results, offering promising potential applications in diagnostics, and data augmentation.
RadGPT: Constructing 3D Image-Text Tumor Datasets
Jan 08, 2025Abstract:With over 85 million CT scans performed annually in the United States, creating tumor-related reports is a challenging and time-consuming task for radiologists. To address this need, we present RadGPT, an Anatomy-Aware Vision-Language AI Agent for generating detailed reports from CT scans. RadGPT first segments tumors, including benign cysts and malignant tumors, and their surrounding anatomical structures, then transforms this information into both structured reports and narrative reports. These reports provide tumor size, shape, location, attenuation, volume, and interactions with surrounding blood vessels and organs. Extensive evaluation on unseen hospitals shows that RadGPT can produce accurate reports, with high sensitivity/specificity for small tumor (<2 cm) detection: 80/73% for liver tumors, 92/78% for kidney tumors, and 77/77% for pancreatic tumors. For large tumors, sensitivity ranges from 89% to 97%. The results significantly surpass the state-of-the-art in abdominal CT report generation. RadGPT generated reports for 17 public datasets. Through radiologist review and refinement, we have ensured the reports' accuracy, and created the first publicly available image-text 3D medical dataset, comprising over 1.8 million text tokens and 2.7 million images from 9,262 CT scans, including 2,947 tumor scans/reports of 8,562 tumor instances. Our reports can: (1) localize tumors in eight liver sub-segments and three pancreatic sub-segments annotated per-voxel; (2) determine pancreatic tumor stage (T1-T4) in 260 reports; and (3) present individual analyses of multiple tumors--rare in human-made reports. Importantly, 948 of the reports are for early-stage tumors.
ScaleMAI: Accelerating the Development of Trusted Datasets and AI Models
Jan 06, 2025

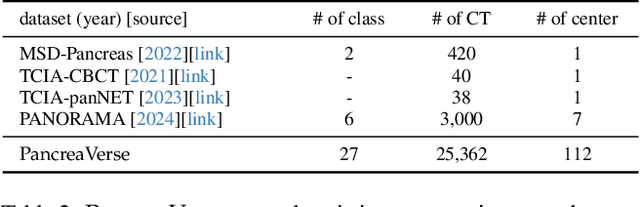

Abstract:Building trusted datasets is critical for transparent and responsible Medical AI (MAI) research, but creating even small, high-quality datasets can take years of effort from multidisciplinary teams. This process often delays AI benefits, as human-centric data creation and AI-centric model development are treated as separate, sequential steps. To overcome this, we propose ScaleMAI, an agent of AI-integrated data curation and annotation, allowing data quality and AI performance to improve in a self-reinforcing cycle and reducing development time from years to months. We adopt pancreatic tumor detection as an example. First, ScaleMAI progressively creates a dataset of 25,362 CT scans, including per-voxel annotations for benign/malignant tumors and 24 anatomical structures. Second, through progressive human-in-the-loop iterations, ScaleMAI provides Flagship AI Model that can approach the proficiency of expert annotators (30-year experience) in detecting pancreatic tumors. Flagship Model significantly outperforms models developed from smaller, fixed-quality datasets, with substantial gains in tumor detection (+14%), segmentation (+5%), and classification (72%) on three prestigious benchmarks. In summary, ScaleMAI transforms the speed, scale, and reliability of medical dataset creation, paving the way for a variety of impactful, data-driven applications.
Text-Driven Tumor Synthesis
Dec 24, 2024



Abstract:Tumor synthesis can generate examples that AI often misses or over-detects, improving AI performance by training on these challenging cases. However, existing synthesis methods, which are typically unconditional -- generating images from random variables -- or conditioned only by tumor shapes, lack controllability over specific tumor characteristics such as texture, heterogeneity, boundaries, and pathology type. As a result, the generated tumors may be overly similar or duplicates of existing training data, failing to effectively address AI's weaknesses. We propose a new text-driven tumor synthesis approach, termed TextoMorph, that provides textual control over tumor characteristics. This is particularly beneficial for examples that confuse the AI the most, such as early tumor detection (increasing Sensitivity by +8.5%), tumor segmentation for precise radiotherapy (increasing DSC by +6.3%), and classification between benign and malignant tumors (improving Sensitivity by +8.2%). By incorporating text mined from radiology reports into the synthesis process, we increase the variability and controllability of the synthetic tumors to target AI's failure cases more precisely. Moreover, TextoMorph uses contrastive learning across different texts and CT scans, significantly reducing dependence on scarce image-report pairs (only 141 pairs used in this study) by leveraging a large corpus of 34,035 radiology reports. Finally, we have developed rigorous tests to evaluate synthetic tumors, including Text-Driven Visual Turing Test and Radiomics Pattern Analysis, showing that our synthetic tumors is realistic and diverse in texture, heterogeneity, boundaries, and pathology.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge