Tianyu Lin
Early and Prediagnostic Detection of Pancreatic Cancer from Computed Tomography
Jan 29, 2026Abstract:Pancreatic ductal adenocarcinoma (PDAC), one of the deadliest solid malignancies, is often detected at a late and inoperable stage. Retrospective reviews of prediagnostic CT scans, when conducted by expert radiologists aware that the patient later developed PDAC, frequently reveal lesions that were previously overlooked. To help detecting these lesions earlier, we developed an automated system named ePAI (early Pancreatic cancer detection with Artificial Intelligence). It was trained on data from 1,598 patients from a single medical center. In the internal test involving 1,009 patients, ePAI achieved an area under the receiver operating characteristic curve (AUC) of 0.939-0.999, a sensitivity of 95.3%, and a specificity of 98.7% for detecting small PDAC less than 2 cm in diameter, precisely localizing PDAC as small as 2 mm. In an external test involving 7,158 patients across 6 centers, ePAI achieved an AUC of 0.918-0.945, a sensitivity of 91.5%, and a specificity of 88.0%, precisely localizing PDAC as small as 5 mm. Importantly, ePAI detected PDACs on prediagnostic CT scans obtained 3 to 36 months before clinical diagnosis that had originally been overlooked by radiologists. It successfully detected and localized PDACs in 75 of 159 patients, with a median lead time of 347 days before clinical diagnosis. Our multi-reader study showed that ePAI significantly outperformed 30 board-certified radiologists by 50.3% (P < 0.05) in sensitivity while maintaining a comparable specificity of 95.4% in detecting PDACs early and prediagnostic. These findings suggest its potential of ePAI as an assistive tool to improve early detection of pancreatic cancer.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Abstract:Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
PanTS: The Pancreatic Tumor Segmentation Dataset
Jul 02, 2025Abstract:PanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
RadFabric: Agentic AI System with Reasoning Capability for Radiology
Jun 17, 2025Abstract:Chest X ray (CXR) imaging remains a critical diagnostic tool for thoracic conditions, but current automated systems face limitations in pathology coverage, diagnostic accuracy, and integration of visual and textual reasoning. To address these gaps, we propose RadFabric, a multi agent, multimodal reasoning framework that unifies visual and textual analysis for comprehensive CXR interpretation. RadFabric is built on the Model Context Protocol (MCP), enabling modularity, interoperability, and scalability for seamless integration of new diagnostic agents. The system employs specialized CXR agents for pathology detection, an Anatomical Interpretation Agent to map visual findings to precise anatomical structures, and a Reasoning Agent powered by large multimodal reasoning models to synthesize visual, anatomical, and clinical data into transparent and evidence based diagnoses. RadFabric achieves significant performance improvements, with near-perfect detection of challenging pathologies like fractures (1.000 accuracy) and superior overall diagnostic accuracy (0.799) compared to traditional systems (0.229 to 0.527). By integrating cross modal feature alignment and preference-driven reasoning, RadFabric advances AI-driven radiology toward transparent, anatomically precise, and clinically actionable CXR analysis.
PGP-SAM: Prototype-Guided Prompt Learning for Efficient Few-Shot Medical Image Segmentation
Jan 12, 2025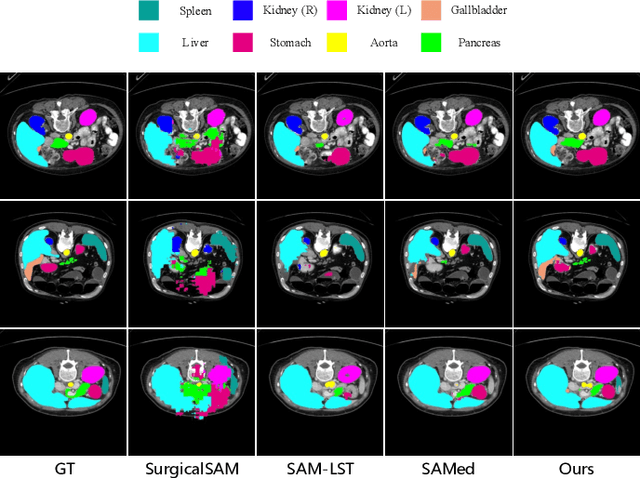



Abstract:The Segment Anything Model (SAM) has demonstrated strong and versatile segmentation capabilities, along with intuitive prompt-based interactions. However, customizing SAM for medical image segmentation requires massive amounts of pixel-level annotations and precise point- or box-based prompt designs. To address these challenges, we introduce PGP-SAM, a novel prototype-based few-shot tuning approach that uses limited samples to replace tedious manual prompts. Our key idea is to leverage inter- and intra-class prototypes to capture class-specific knowledge and relationships. We propose two main components: (1) a plug-and-play contextual modulation module that integrates multi-scale information, and (2) a class-guided cross-attention mechanism that fuses prototypes and features for automatic prompt generation. Experiments on a public multi-organ dataset and a private ventricle dataset demonstrate that PGP-SAM achieves superior mean Dice scores compared with existing prompt-free SAM variants, while using only 10\% of the 2D slices.
ScaleMAI: Accelerating the Development of Trusted Datasets and AI Models
Jan 06, 2025

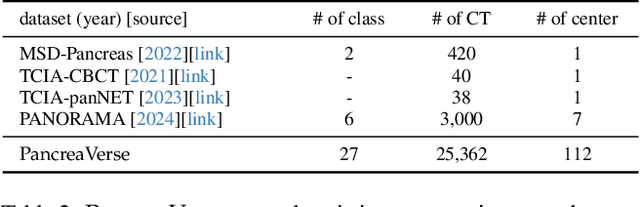

Abstract:Building trusted datasets is critical for transparent and responsible Medical AI (MAI) research, but creating even small, high-quality datasets can take years of effort from multidisciplinary teams. This process often delays AI benefits, as human-centric data creation and AI-centric model development are treated as separate, sequential steps. To overcome this, we propose ScaleMAI, an agent of AI-integrated data curation and annotation, allowing data quality and AI performance to improve in a self-reinforcing cycle and reducing development time from years to months. We adopt pancreatic tumor detection as an example. First, ScaleMAI progressively creates a dataset of 25,362 CT scans, including per-voxel annotations for benign/malignant tumors and 24 anatomical structures. Second, through progressive human-in-the-loop iterations, ScaleMAI provides Flagship AI Model that can approach the proficiency of expert annotators (30-year experience) in detecting pancreatic tumors. Flagship Model significantly outperforms models developed from smaller, fixed-quality datasets, with substantial gains in tumor detection (+14%), segmentation (+5%), and classification (72%) on three prestigious benchmarks. In summary, ScaleMAI transforms the speed, scale, and reliability of medical dataset creation, paving the way for a variety of impactful, data-driven applications.
Stable Diffusion Segmentation for Biomedical Images with Single-step Reverse Process
Jun 27, 2024



Abstract:Diffusion models have demonstrated their effectiveness across various generative tasks. However, when applied to medical image segmentation, these models encounter several challenges, including significant resource and time requirements. They also necessitate a multi-step reverse process and multiple samples to produce reliable predictions. To address these challenges, we introduce the first latent diffusion segmentation model, named SDSeg, built upon stable diffusion (SD). SDSeg incorporates a straightforward latent estimation strategy to facilitate a single-step reverse process and utilizes latent fusion concatenation to remove the necessity for multiple samples. Extensive experiments indicate that SDSeg surpasses existing state-of-the-art methods on five benchmark datasets featuring diverse imaging modalities. Remarkably, SDSeg is capable of generating stable predictions with a solitary reverse step and sample, epitomizing the model's stability as implied by its name. The code is available at https://github.com/lin-tianyu/Stable-Diffusion-Seg
Dimension Independent Mixup for Hard Negative Sample in Collaborative Filtering
Jun 28, 2023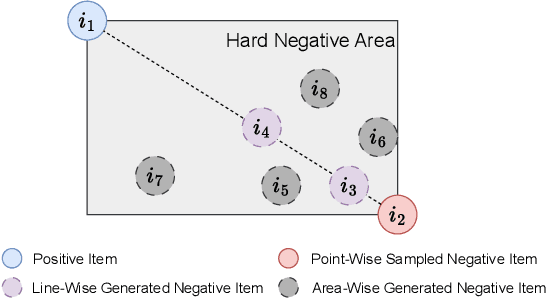



Abstract:Collaborative filtering (CF) is a widely employed technique that predicts user preferences based on past interactions. Negative sampling plays a vital role in training CF-based models with implicit feedback. In this paper, we propose a novel perspective based on the sampling area to revisit existing sampling methods. We point out that current sampling methods mainly focus on Point-wise or Line-wise sampling, lacking flexibility and leaving a significant portion of the hard sampling area un-explored. To address this limitation, we propose Dimension Independent Mixup for Hard Negative Sampling (DINS), which is the first Area-wise sampling method for training CF-based models. DINS comprises three modules: Hard Boundary Definition, Dimension Independent Mixup, and Multi-hop Pooling. Experiments with real-world datasets on both matrix factorization and graph-based models demonstrate that DINS outperforms other negative sampling methods, establishing its effectiveness and superiority. Our work contributes a new perspective, introduces Area-wise sampling, and presents DINS as a novel approach that achieves state-of-the-art performance for negative sampling. Our implementations are available in PyTorch.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge