Xiangzhu Zeng
PGP-SAM: Prototype-Guided Prompt Learning for Efficient Few-Shot Medical Image Segmentation
Jan 12, 2025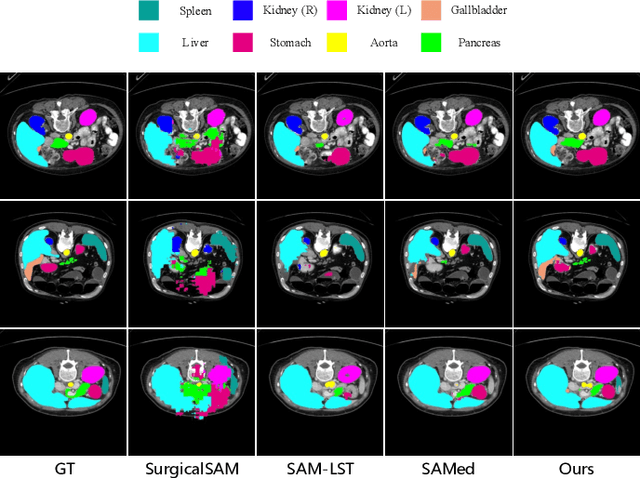



Abstract:The Segment Anything Model (SAM) has demonstrated strong and versatile segmentation capabilities, along with intuitive prompt-based interactions. However, customizing SAM for medical image segmentation requires massive amounts of pixel-level annotations and precise point- or box-based prompt designs. To address these challenges, we introduce PGP-SAM, a novel prototype-based few-shot tuning approach that uses limited samples to replace tedious manual prompts. Our key idea is to leverage inter- and intra-class prototypes to capture class-specific knowledge and relationships. We propose two main components: (1) a plug-and-play contextual modulation module that integrates multi-scale information, and (2) a class-guided cross-attention mechanism that fuses prototypes and features for automatic prompt generation. Experiments on a public multi-organ dataset and a private ventricle dataset demonstrate that PGP-SAM achieves superior mean Dice scores compared with existing prompt-free SAM variants, while using only 10\% of the 2D slices.
CarveMix: A Simple Data Augmentation Method for Brain Lesion Segmentation
Aug 17, 2021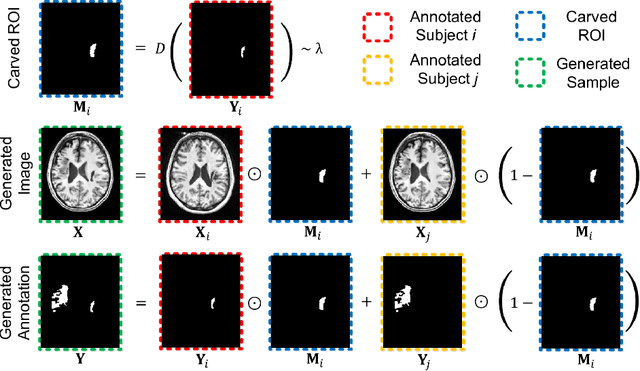
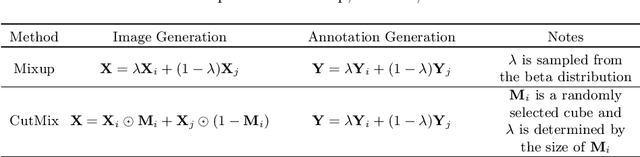
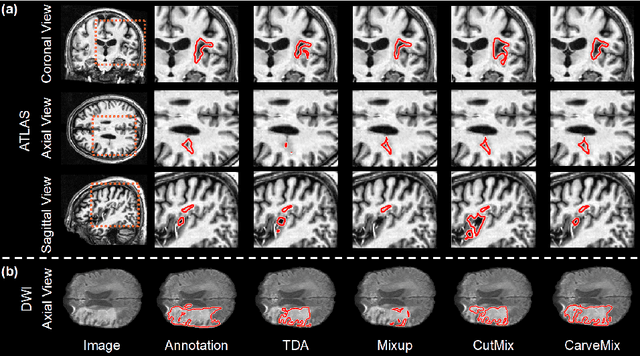
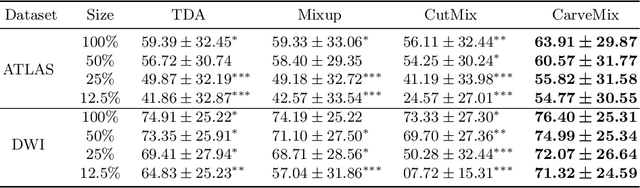
Abstract:Brain lesion segmentation provides a valuable tool for clinical diagnosis, and convolutional neural networks (CNNs) have achieved unprecedented success in the task. Data augmentation is a widely used strategy that improves the training of CNNs, and the design of the augmentation method for brain lesion segmentation is still an open problem. In this work, we propose a simple data augmentation approach, dubbed as CarveMix, for CNN-based brain lesion segmentation. Like other "mix"-based methods, such as Mixup and CutMix, CarveMix stochastically combines two existing labeled images to generate new labeled samples. Yet, unlike these augmentation strategies based on image combination, CarveMix is lesion-aware, where the combination is performed with an attention on the lesions and a proper annotation is created for the generated image. Specifically, from one labeled image we carve a region of interest (ROI) according to the lesion location and geometry, and the size of the ROI is sampled from a probability distribution. The carved ROI then replaces the corresponding voxels in a second labeled image, and the annotation of the second image is replaced accordingly as well. In this way, we generate new labeled images for network training and the lesion information is preserved. To evaluate the proposed method, experiments were performed on two brain lesion datasets. The results show that our method improves the segmentation accuracy compared with other simple data augmentation approaches.
Semi-Supervised Brain Lesion Segmentation with an Adapted Mean Teacher Model
Mar 04, 2019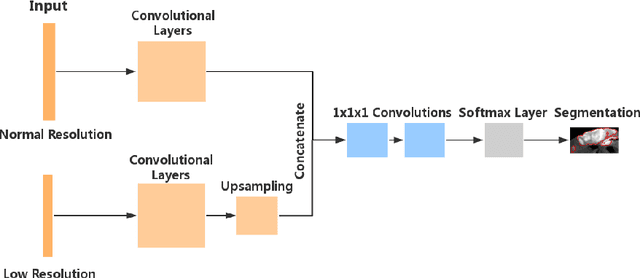
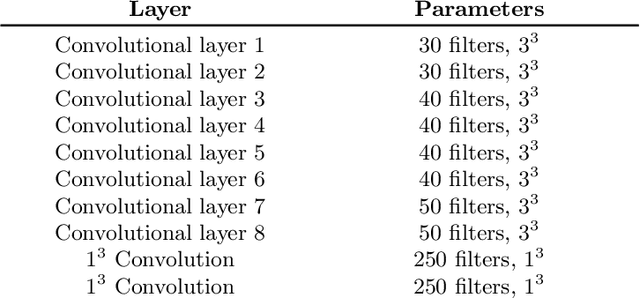
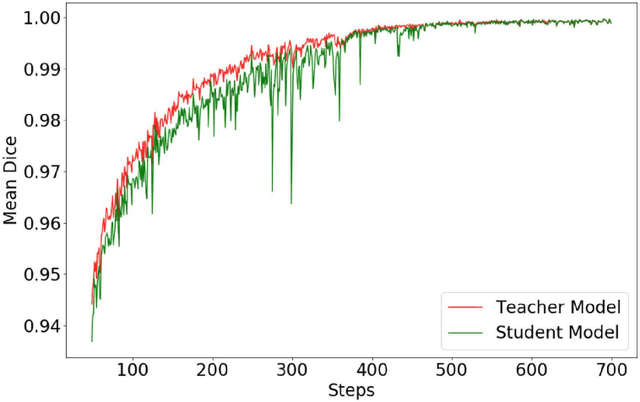
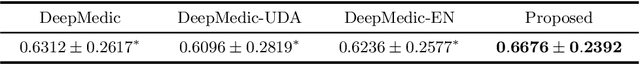
Abstract:Automated brain lesion segmentation provides valuable information for the analysis and intervention of patients. In particular, methods based on convolutional neural networks (CNNs) have achieved state-of-the-art segmentation performance. However, CNNs usually require a decent amount of annotated data, which may be costly and time-consuming to obtain. Since unannotated data is generally abundant, it is desirable to use unannotated data to improve the segmentation performance for CNNs when limited annotated data is available. In this work, we propose a semi-supervised learning (SSL) approach to brain lesion segmentation, where unannotated data is incorporated into the training of CNNs. We adapt the mean teacher model, which is originally developed for SSL-based image classification, for brain lesion segmentation. Assuming that the network should produce consistent outputs for similar inputs, a loss of segmentation consistency is designed and integrated into a self-ensembling framework. Specifically, we build a student model and a teacher model, which share the same CNN architecture for segmentation. The student and teacher models are updated alternately. At each step, the student model learns from the teacher model by minimizing the weighted sum of the segmentation loss computed from annotated data and the segmentation consistency loss between the teacher and student models computed from unannotated data. Then, the teacher model is updated by combining the updated student model with the historical information of teacher models using an exponential moving average strategy. For demonstration, the proposed approach was evaluated on ischemic stroke lesion segmentation, where it improves stroke lesion segmentation with the incorporation of unannotated data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge