Zhiwen Liu
The Second Monocular Depth Estimation Challenge
Apr 26, 2023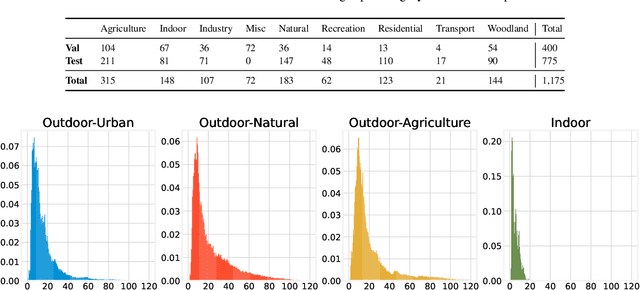
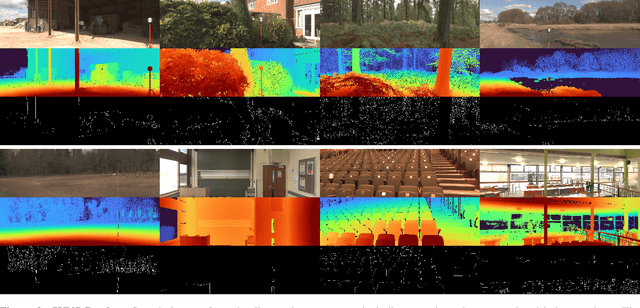
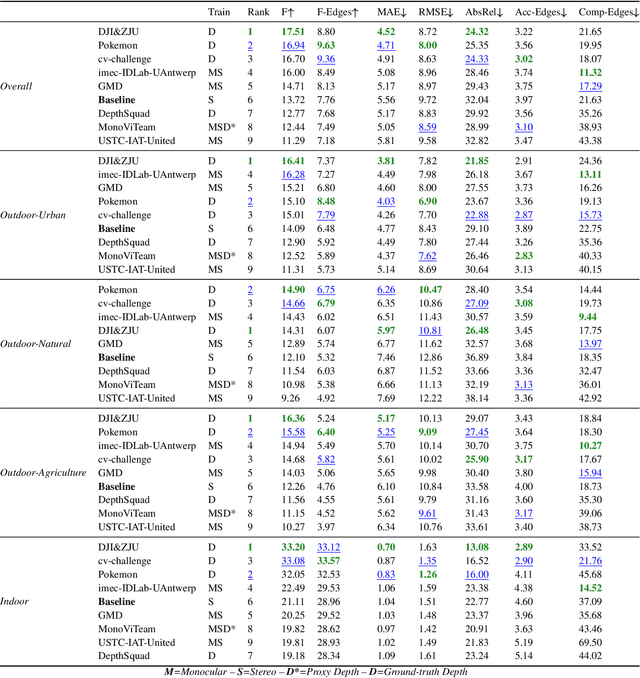
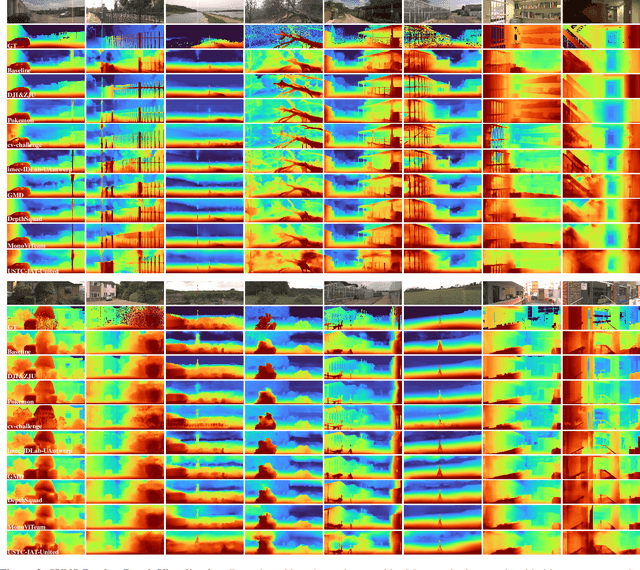
Abstract:This paper discusses the results for the second edition of the Monocular Depth Estimation Challenge (MDEC). This edition was open to methods using any form of supervision, including fully-supervised, self-supervised, multi-task or proxy depth. The challenge was based around the SYNS-Patches dataset, which features a wide diversity of environments with high-quality dense ground-truth. This includes complex natural environments, e.g. forests or fields, which are greatly underrepresented in current benchmarks. The challenge received eight unique submissions that outperformed the provided SotA baseline on any of the pointcloud- or image-based metrics. The top supervised submission improved relative F-Score by 27.62%, while the top self-supervised improved it by 16.61%. Supervised submissions generally leveraged large collections of datasets to improve data diversity. Self-supervised submissions instead updated the network architecture and pretrained backbones. These results represent a significant progress in the field, while highlighting avenues for future research, such as reducing interpolation artifacts at depth boundaries, improving self-supervised indoor performance and overall natural image accuracy.
Positive-unlabeled learning for binary and multi-class cell detection in histopathology images with incomplete annotations
Feb 16, 2023Abstract:Cell detection in histopathology images is of great interest to clinical practice and research, and convolutional neural networks (CNNs) have achieved remarkable cell detection results. Typically, to train CNN-based cell detection models, every positive instance in the training images needs to be annotated, and instances that are not labeled as positive are considered negative samples. However, manual cell annotation is complicated due to the large number and diversity of cells, and it can be difficult to ensure the annotation of every positive instance. In many cases, only incomplete annotations are available, where some of the positive instances are annotated and the others are not, and the classification loss term for negative samples in typical network training becomes incorrect. In this work, to address this problem of incomplete annotations, we propose to reformulate the training of the detection network as a positive-unlabeled learning problem. Since the instances in unannotated regions can be either positive or negative, they have unknown labels. Using the samples with unknown labels and the positively labeled samples, we first derive an approximation of the classification loss term corresponding to negative samples for binary cell detection, and based on this approximation we further extend the proposed framework to multi-class cell detection. For evaluation, experiments were performed on four publicly available datasets. The experimental results show that our method improves the performance of cell detection in histopathology images given incomplete annotations for network training.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2022:027. arXiv admin note: text overlap with arXiv:2106.15918
CarveMix: A Simple Data Augmentation Method for Brain Lesion Segmentation
Aug 17, 2021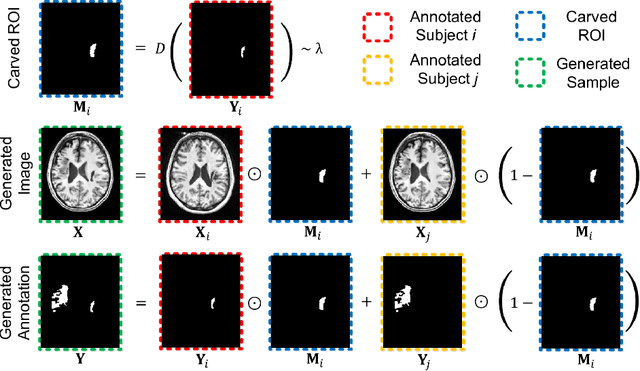
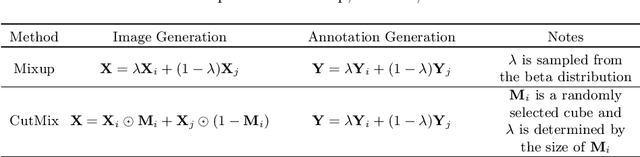
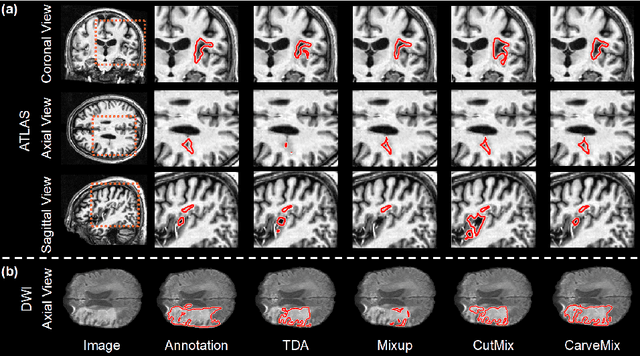
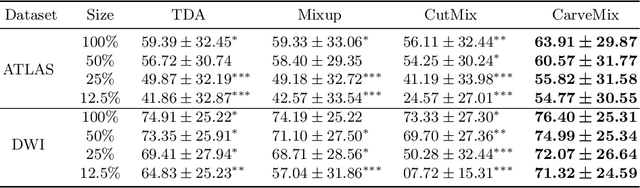
Abstract:Brain lesion segmentation provides a valuable tool for clinical diagnosis, and convolutional neural networks (CNNs) have achieved unprecedented success in the task. Data augmentation is a widely used strategy that improves the training of CNNs, and the design of the augmentation method for brain lesion segmentation is still an open problem. In this work, we propose a simple data augmentation approach, dubbed as CarveMix, for CNN-based brain lesion segmentation. Like other "mix"-based methods, such as Mixup and CutMix, CarveMix stochastically combines two existing labeled images to generate new labeled samples. Yet, unlike these augmentation strategies based on image combination, CarveMix is lesion-aware, where the combination is performed with an attention on the lesions and a proper annotation is created for the generated image. Specifically, from one labeled image we carve a region of interest (ROI) according to the lesion location and geometry, and the size of the ROI is sampled from a probability distribution. The carved ROI then replaces the corresponding voxels in a second labeled image, and the annotation of the second image is replaced accordingly as well. In this way, we generate new labeled images for network training and the lesion information is preserved. To evaluate the proposed method, experiments were performed on two brain lesion datasets. The results show that our method improves the segmentation accuracy compared with other simple data augmentation approaches.
Positive-unlabeled Learning for Cell Detection in Histopathology Images with Incomplete Annotations
Jun 30, 2021

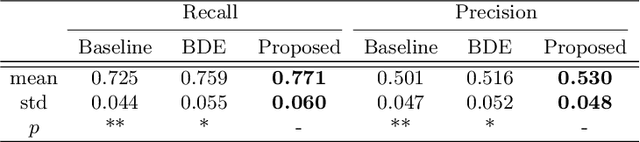
Abstract:Cell detection in histopathology images is of great value in clinical practice. \textit{Convolutional neural networks} (CNNs) have been applied to cell detection to improve the detection accuracy, where cell annotations are required for network training. However, due to the variety and large number of cells, complete annotations that include every cell of interest in the training images can be challenging. Usually, incomplete annotations can be achieved, where positive labeling results are carefully examined to ensure their reliability but there can be other positive instances, i.e., cells of interest, that are not included in the annotations. This annotation strategy leads to a lack of knowledge about true negative samples. Most existing methods simply treat instances that are not labeled as positive as truly negative during network training, which can adversely affect the network performance. In this work, to address the problem of incomplete annotations, we formulate the training of detection networks as a positive-unlabeled learning problem. Specifically, the classification loss in network training is revised to take into account incomplete annotations, where the terms corresponding to negative samples are approximated with the true positive samples and the other samples of which the labels are unknown. To evaluate the proposed method, experiments were performed on a publicly available dataset for mitosis detection in breast cancer cells, and the experimental results show that our method improves the performance of cell detection given incomplete annotations for training.
Knowledge Transfer between Datasets for Learning-based Tissue Microstructure Estimation
Oct 24, 2019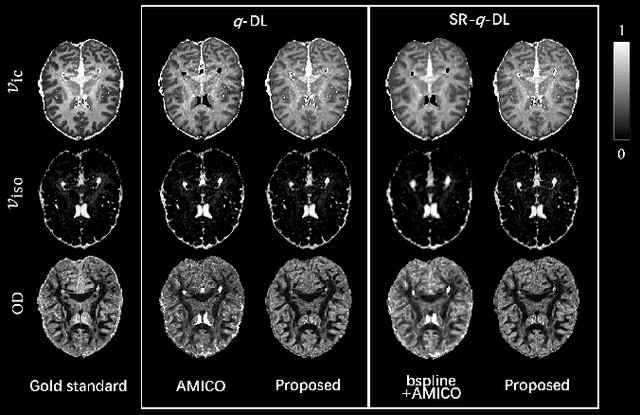
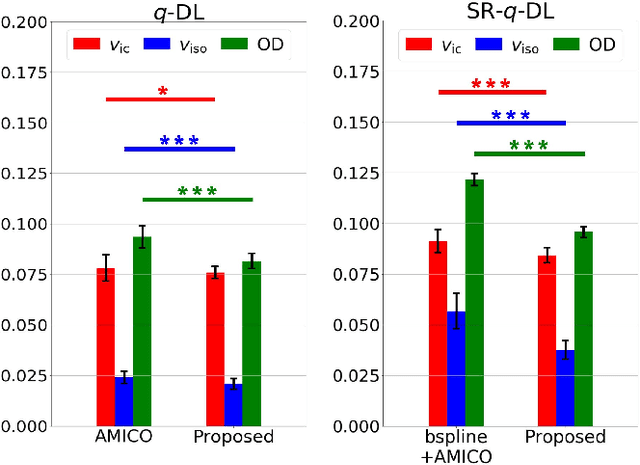
Abstract:Learning-based approaches, especially those based on deep networks, have enabled high-quality estimation of tissue microstructure from low-quality diffusion magnetic resonance imaging (dMRI) scans, which are acquired with a limited number of diffusion gradients and a relatively poor spatial resolution. These learning-based approaches to tissue microstructure estimation require acquisitions of training dMRI scans with high-quality diffusion signals, which are densely sampled in the q-space and have a high spatial resolution. However, the acquisition of training scans may not be available for all datasets. Therefore, we explore knowledge transfer between different dMRI datasets so that learning-based tissue microstructure estimation can be applied for datasets where training scans are not acquired. Specifically, for a target dataset of interest, where only low-quality diffusion signals are acquired without training scans, we exploit the information in a source dMRI dataset acquired with high-quality diffusion signals. We interpolate the diffusion signals in the source dataset in the q-space using a dictionary-based signal representation, so that the interpolated signals match the acquisition scheme of the target dataset. Then, the interpolated signals are used together with the high-quality tissue microstructure computed from the source dataset to train deep networks that perform tissue microstructure estimation for the target dataset. Experiments were performed on brain dMRI scans with low-quality diffusion signals, where the benefit of the proposed strategy is demonstrated.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge