Yaou Liu
A Foundation Model for Brain Lesion Segmentation with Mixture of Modality Experts
May 16, 2024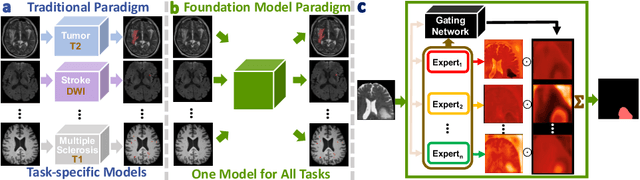
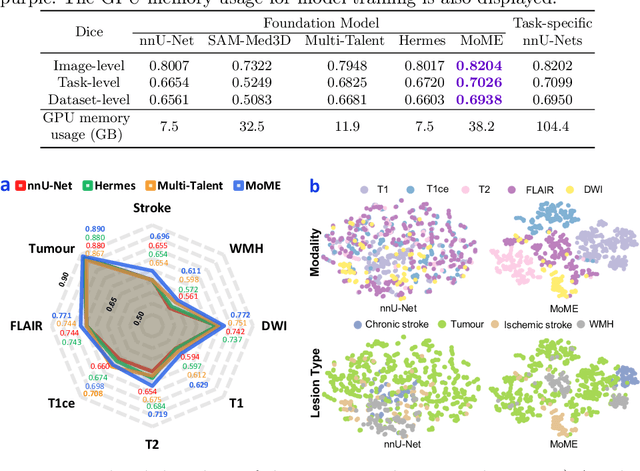

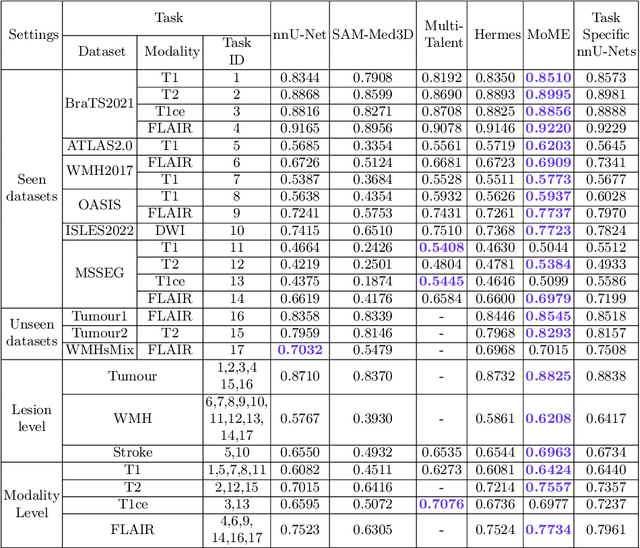
Abstract:Brain lesion segmentation plays an essential role in neurological research and diagnosis. As brain lesions can be caused by various pathological alterations, different types of brain lesions tend to manifest with different characteristics on different imaging modalities. Due to this complexity, brain lesion segmentation methods are often developed in a task-specific manner. A specific segmentation model is developed for a particular lesion type and imaging modality. However, the use of task-specific models requires predetermination of the lesion type and imaging modality, which complicates their deployment in real-world scenarios. In this work, we propose a universal foundation model for 3D brain lesion segmentation, which can automatically segment different types of brain lesions for input data of various imaging modalities. We formulate a novel Mixture of Modality Experts (MoME) framework with multiple expert networks attending to different imaging modalities. A hierarchical gating network combines the expert predictions and fosters expertise collaboration. Furthermore, we introduce a curriculum learning strategy during training to avoid the degeneration of each expert network and preserve their specialization. We evaluated the proposed method on nine brain lesion datasets, encompassing five imaging modalities and eight lesion types. The results show that our model outperforms state-of-the-art universal models and provides promising generalization to unseen datasets.
Pathology-genomic fusion via biologically informed cross-modality graph learning for survival analysis
Apr 11, 2024



Abstract:The diagnosis and prognosis of cancer are typically based on multi-modal clinical data, including histology images and genomic data, due to the complex pathogenesis and high heterogeneity. Despite the advancements in digital pathology and high-throughput genome sequencing, establishing effective multi-modal fusion models for survival prediction and revealing the potential association between histopathology and transcriptomics remains challenging. In this paper, we propose Pathology-Genome Heterogeneous Graph (PGHG) that integrates whole slide images (WSI) and bulk RNA-Seq expression data with heterogeneous graph neural network for cancer survival analysis. The PGHG consists of biological knowledge-guided representation learning network and pathology-genome heterogeneous graph. The representation learning network utilizes the biological prior knowledge of intra-modal and inter-modal data associations to guide the feature extraction. The node features of each modality are updated through attention-based graph learning strategy. Unimodal features and bi-modal fused features are extracted via attention pooling module and then used for survival prediction. We evaluate the model on low-grade gliomas, glioblastoma, and kidney renal papillary cell carcinoma datasets from the Cancer Genome Atlas (TCGA) and the First Affiliated Hospital of Zhengzhou University (FAHZU). Extensive experimental results demonstrate that the proposed method outperforms both unimodal and other multi-modal fusion models. For demonstrating the model interpretability, we also visualize the attention heatmap of pathological images and utilize integrated gradient algorithm to identify important tissue structure, biological pathways and key genes.
Unsupervised Brain Tumor Segmentation with Image-based Prompts
Apr 04, 2023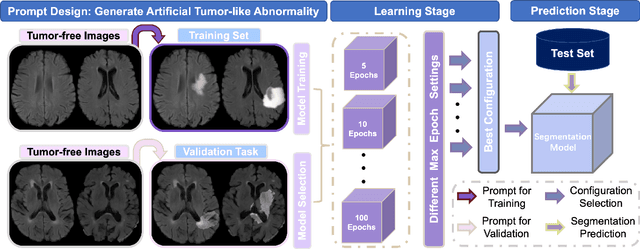
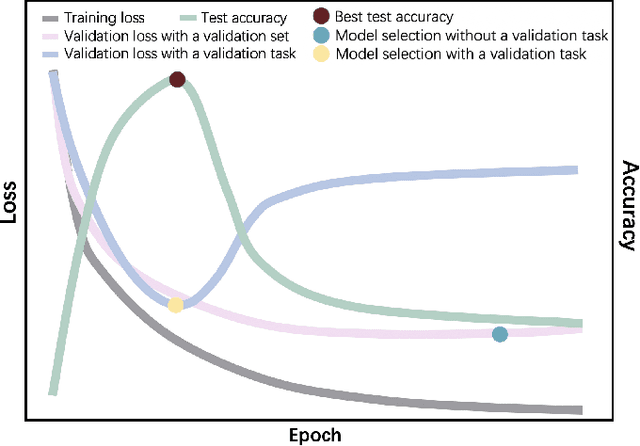
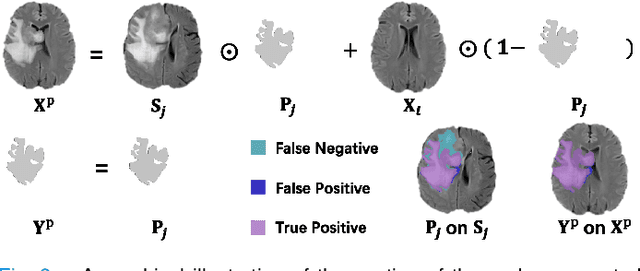
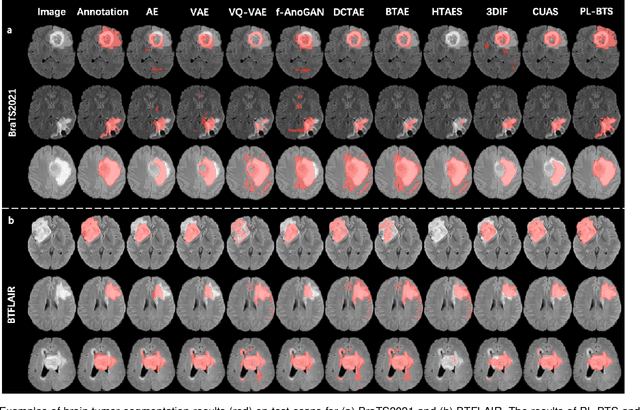
Abstract:Automated brain tumor segmentation based on deep learning (DL) has achieved promising performance. However, it generally relies on annotated images for model training, which is not always feasible in clinical settings. Therefore, the development of unsupervised DL-based brain tumor segmentation approaches without expert annotations is desired. Motivated by the success of prompt learning (PL) in natural language processing, we propose an approach to unsupervised brain tumor segmentation by designing image-based prompts that allow indication of brain tumors, and this approach is dubbed as PL-based Brain Tumor Segmentation (PL-BTS). Specifically, instead of directly training a model for brain tumor segmentation with a large amount of annotated data, we seek to train a model that can answer the question: is a voxel in the input image associated with tumor-like hyper-/hypo-intensity? Such a model can be trained by artificially generating tumor-like hyper-/hypo-intensity on images without tumors with hand-crafted designs. Since the hand-crafted designs may be too simplistic to represent all kinds of real tumors, the trained model may overfit the simplistic hand-crafted task rather than actually answer the question of abnormality. To address this problem, we propose the use of a validation task, where we generate a different hand-crafted task to monitor overfitting. In addition, we propose PL-BTS+ that further improves PL-BTS by exploiting unannotated images with brain tumors. Compared with competing unsupervised methods, the proposed method has achieved marked improvements on both public and in-house datasets, and we have also demonstrated its possible extension to other brain lesion segmentation tasks.
One-Shot Segmentation of Novel White Matter Tracts via Extensive Data Augmentation
Mar 13, 2023Abstract:Deep learning based methods have achieved state-of-the-art performance for automated white matter (WM) tract segmentation. In these methods, the segmentation model needs to be trained with a large number of manually annotated scans, which can be accumulated throughout time. When novel WM tracts, i.e., tracts not included in the existing annotated WM tracts, are to be segmented, additional annotations of these novel WM tracts need to be collected. Since tract annotation is time-consuming and costly, it is desirable to make only a few annotations of novel WM tracts for training the segmentation model, and previous work has addressed this problem by transferring the knowledge learned for segmenting existing WM tracts to the segmentation of novel WM tracts. However, accurate segmentation of novel WM tracts can still be challenging in the one-shot setting, where only one scan is annotated for the novel WM tracts. In this work, we explore the problem of one-shot segmentation of novel WM tracts. Since in the one-shot setting the annotated training data is extremely scarce, based on the existing knowledge transfer framework, we propose to further perform extensive data augmentation for the single annotated scan, where synthetic annotated training data is produced. We have designed several different strategies that mask out regions in the single annotated scan for data augmentation. Our method was evaluated on public and in-house datasets. The experimental results show that our method improves the accuracy of one-shot segmentation of novel WM tracts.
Positive-unlabeled learning for binary and multi-class cell detection in histopathology images with incomplete annotations
Feb 16, 2023Abstract:Cell detection in histopathology images is of great interest to clinical practice and research, and convolutional neural networks (CNNs) have achieved remarkable cell detection results. Typically, to train CNN-based cell detection models, every positive instance in the training images needs to be annotated, and instances that are not labeled as positive are considered negative samples. However, manual cell annotation is complicated due to the large number and diversity of cells, and it can be difficult to ensure the annotation of every positive instance. In many cases, only incomplete annotations are available, where some of the positive instances are annotated and the others are not, and the classification loss term for negative samples in typical network training becomes incorrect. In this work, to address this problem of incomplete annotations, we propose to reformulate the training of the detection network as a positive-unlabeled learning problem. Since the instances in unannotated regions can be either positive or negative, they have unknown labels. Using the samples with unknown labels and the positively labeled samples, we first derive an approximation of the classification loss term corresponding to negative samples for binary cell detection, and based on this approximation we further extend the proposed framework to multi-class cell detection. For evaluation, experiments were performed on four publicly available datasets. The experimental results show that our method improves the performance of cell detection in histopathology images given incomplete annotations for network training.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2022:027. arXiv admin note: text overlap with arXiv:2106.15918
Benefits of Linear Conditioning for Segmentation using Metadata
Feb 18, 2021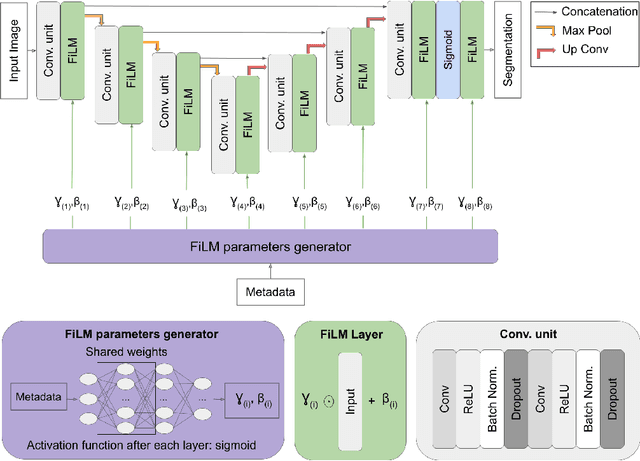

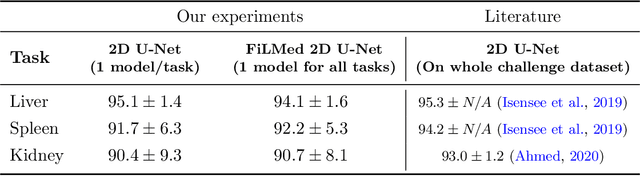

Abstract:Medical images are often accompanied by metadata describing the image (vendor, acquisition parameters) and the patient (disease type or severity, demographics, genomics). This metadata is usually disregarded by image segmentation methods. In this work, we adapt a linear conditioning method called FiLM (Feature-wise Linear Modulation) for image segmentation tasks. This FiLM adaptation enables integrating metadata into segmentation models for better performance. We observed an average Dice score increase of 5.1% on spinal cord tumor segmentation when incorporating the tumor type with FiLM. The metadata modulates the segmentation process through low-cost affine transformations applied on feature maps which can be included in any neural network's architecture. Additionally, we assess the relevance of segmentation FiLM layers for tackling common challenges in medical imaging: training with limited or unbalanced number of annotated data, multi-class training with missing segmentations, and model adaptation to multiple tasks. Our results demonstrated the following benefits of FiLM for segmentation: FiLMed U-Net was robust to missing labels and reached higher Dice scores with few labels (up to 16.7%) compared to single-task U-Net. The code is open-source and available at www.ivadomed.org.
Multiclass Spinal Cord Tumor Segmentation on MRI with Deep Learning
Jan 14, 2021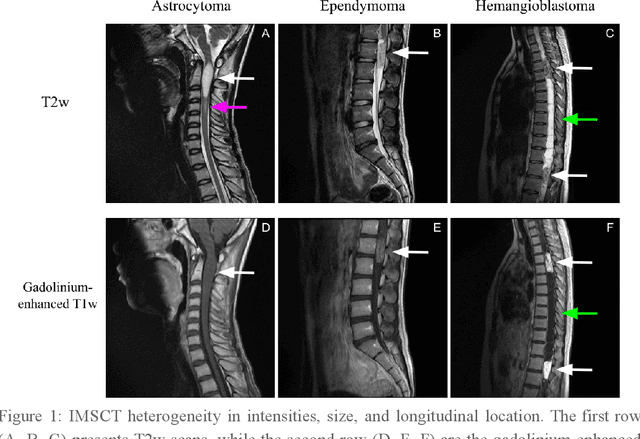
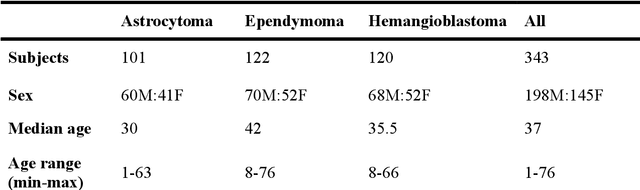
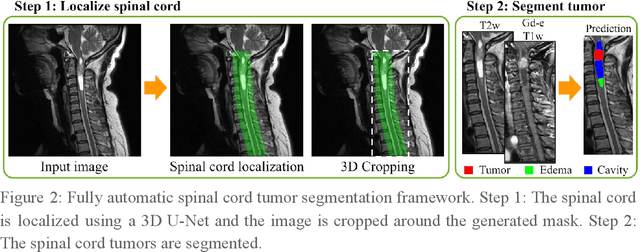
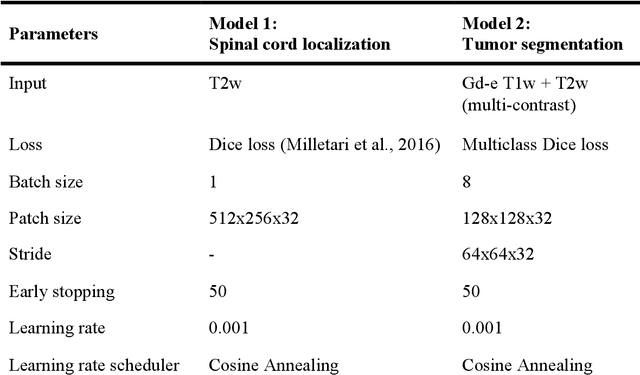
Abstract:Spinal cord tumors lead to neurological morbidity and mortality. Being able to obtain morphometric quantification (size, location, growth rate) of the tumor, edema, and cavity can result in improved monitoring and treatment planning. Such quantification requires the segmentation of these structures into three separate classes. However, manual segmentation of 3-dimensional structures is time-consuming and tedious, motivating the development of automated methods. Here, we tailor a model adapted to the spinal cord tumor segmentation task. Data were obtained from 343 patients using gadolinium-enhanced T1-weighted and T2-weighted MRI scans with cervical, thoracic, and/or lumbar coverage. The dataset includes the three most common intramedullary spinal cord tumor types: astrocytomas, ependymomas, and hemangioblastomas. The proposed approach is a cascaded architecture with U-Net-based models that segments tumors in a two-stage process: locate and label. The model first finds the spinal cord and generates bounding box coordinates. The images are cropped according to this output, leading to a reduced field of view, which mitigates class imbalance. The tumor is then segmented. The segmentation of the tumor, cavity, and edema (as a single class) reached 76.7 $\pm$ 1.5% of Dice score and the segmentation of tumors alone reached 61.8 $\pm$ 4.0% Dice score. The true positive detection rate was above 87% for tumor, edema, and cavity. To the best of our knowledge, this is the first fully automatic deep learning model for spinal cord tumor segmentation. The multiclass segmentation pipeline is available in the Spinal Cord Toolbox (https://spinalcordtoolbox.com/). It can be run with custom data on a regular computer within seconds.
Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks
Sep 11, 2018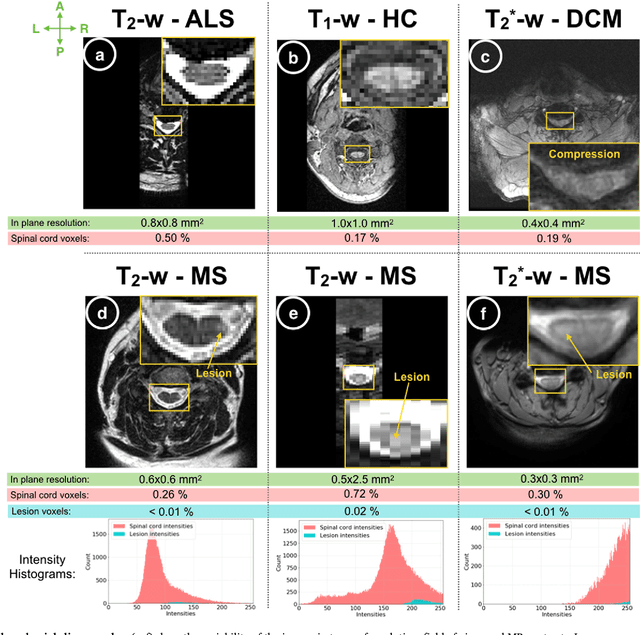
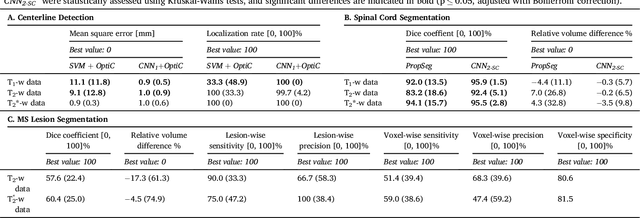
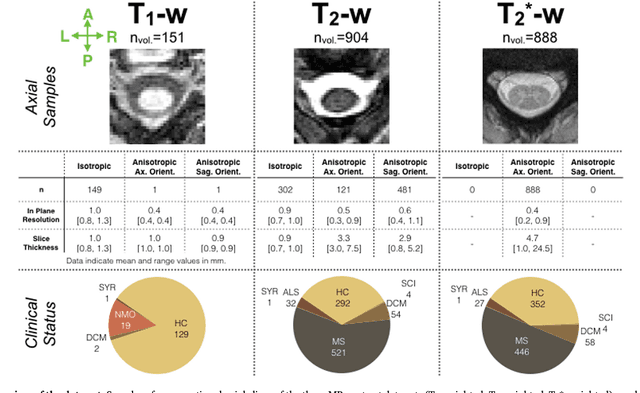
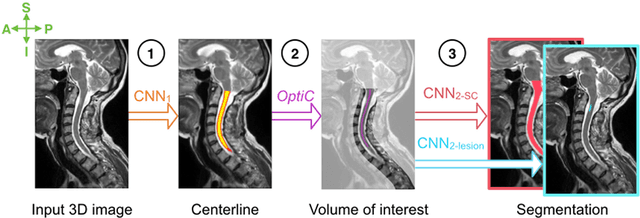
Abstract:The spinal cord is frequently affected by atrophy and/or lesions in multiple sclerosis (MS) patients. Segmentation of the spinal cord and lesions from MRI data provides measures of damage, which are key criteria for the diagnosis, prognosis, and longitudinal monitoring in MS. Automating this operation eliminates inter-rater variability and increases the efficiency of large-throughput analysis pipelines. Robust and reliable segmentation across multi-site spinal cord data is challenging because of the large variability related to acquisition parameters and image artifacts. The goal of this study was to develop a fully-automatic framework, robust to variability in both image parameters and clinical condition, for segmentation of the spinal cord and intramedullary MS lesions from conventional MRI data. Scans of 1,042 subjects (459 healthy controls, 471 MS patients, and 112 with other spinal pathologies) were included in this multi-site study (n=30). Data spanned three contrasts (T1-, T2-, and T2*-weighted) for a total of 1,943 volumes. The proposed cord and lesion automatic segmentation approach is based on a sequence of two Convolutional Neural Networks (CNNs). To deal with the very small proportion of spinal cord and/or lesion voxels compared to the rest of the volume, a first CNN with 2D dilated convolutions detects the spinal cord centerline, followed by a second CNN with 3D convolutions that segments the spinal cord and/or lesions. When compared against manual segmentation, our CNN-based approach showed a median Dice of 95% vs. 88% for PropSeg, a state-of-the-art spinal cord segmentation method. Regarding lesion segmentation on MS data, our framework provided a Dice of 60%, a relative volume difference of -15%, and a lesion-wise detection sensitivity and precision of 83% and 77%, respectively. The proposed framework is open-source and readily available in the Spinal Cord Toolbox.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge