Hairong Zheng
HAVIR: HierArchical Vision to Image Reconstruction using CLIP-Guided Versatile Diffusion
Jun 06, 2025Abstract:Reconstructing visual information from brain activity bridges the gap between neuroscience and computer vision. Even though progress has been made in decoding images from fMRI using generative models, a challenge remains in accurately recovering highly complex visual stimuli. This difficulty stems from their elemental density and diversity, sophisticated spatial structures, and multifaceted semantic information. To address these challenges, we propose HAVIR that contains two adapters: (1) The AutoKL Adapter transforms fMRI voxels into a latent diffusion prior, capturing topological structures; (2) The CLIP Adapter converts the voxels to CLIP text and image embeddings, containing semantic information. These complementary representations are fused by Versatile Diffusion to generate the final reconstructed image. To extract the most essential semantic information from complex scenarios, the CLIP Adapter is trained with text captions describing the visual stimuli and their corresponding semantic images synthesized from these captions. The experimental results demonstrate that HAVIR effectively reconstructs both structural features and semantic information of visual stimuli even in complex scenarios, outperforming existing models.
Patch-based Reconstruction for Unsupervised Dynamic MRI using Learnable Tensor Function with Implicit Neural Representation
May 28, 2025Abstract:Dynamic MRI plays a vital role in clinical practice by capturing both spatial details and dynamic motion, but its high spatiotemporal resolution is often limited by long scan times. Deep learning (DL)-based methods have shown promising performance in accelerating dynamic MRI. However, most existing algorithms rely on large fully-sampled datasets for training, which are difficult to acquire. Recently, implicit neural representation (INR) has emerged as a powerful scan-specific paradigm for accelerated MRI, which models signals as a continuous function over spatiotemporal coordinates. Although this approach achieves efficient continuous modeling of dynamic images and robust reconstruction, it faces challenges in recovering fine details and increasing computational demands for high dimensional data representation. To enhance both efficiency and reconstruction quality, we propose TenF-INR, a novel patch-based unsupervised framework that employs INR to model bases of tensor decomposition, enabling efficient and accurate modeling of dynamic MR images with learnable tensor functions. By exploiting strong correlations in similar spatial image patches and in the temporal direction, TenF-INR enforces multidimensional low-rankness and implements patch-based reconstruction with the benefits of continuous modeling. We compare TenF-INR with state-of-the-art methods, including supervised DL methods and unsupervised approaches. Experimental results demonstrate that TenF-INR achieves high acceleration factors up to 21, outperforming all comparison methods in image quality, temporal fidelity, and quantitative metrics, even surpassing the supervised methods.
Cross-Sequence Semi-Supervised Learning for Multi-Parametric MRI-Based Visual Pathway Delineation
May 26, 2025Abstract:Accurately delineating the visual pathway (VP) is crucial for understanding the human visual system and diagnosing related disorders. Exploring multi-parametric MR imaging data has been identified as an important way to delineate VP. However, due to the complex cross-sequence relationships, existing methods cannot effectively model the complementary information from different MRI sequences. In addition, these existing methods heavily rely on large training data with labels, which is labor-intensive and time-consuming to obtain. In this work, we propose a novel semi-supervised multi-parametric feature decomposition framework for VP delineation. Specifically, a correlation-constrained feature decomposition (CFD) is designed to handle the complex cross-sequence relationships by capturing the unique characteristics of each MRI sequence and easing the multi-parametric information fusion process. Furthermore, a consistency-based sample enhancement (CSE) module is developed to address the limited labeled data issue, by generating and promoting meaningful edge information from unlabeled data. We validate our framework using two public datasets, and one in-house Multi-Shell Diffusion MRI (MDM) dataset. Experimental results demonstrate the superiority of our approach in terms of delineation performance when compared to seven state-of-the-art approaches.
Diff5T: Benchmarking Human Brain Diffusion MRI with an Extensive 5.0 Tesla K-Space and Spatial Dataset
Dec 09, 2024

Abstract:Diffusion magnetic resonance imaging (dMRI) provides critical insights into the microstructural and connectional organization of the human brain. However, the availability of high-field, open-access datasets that include raw k-space data for advanced research remains limited. To address this gap, we introduce Diff5T, a first comprehensive 5.0 Tesla diffusion MRI dataset focusing on the human brain. This dataset includes raw k-space data and reconstructed diffusion images, acquired using a variety of imaging protocols. Diff5T is designed to support the development and benchmarking of innovative methods in artifact correction, image reconstruction, image preprocessing, diffusion modelling and tractography. The dataset features a wide range of diffusion parameters, including multiple b-values and gradient directions, allowing extensive research applications in studying human brain microstructure and connectivity. With its emphasis on open accessibility and detailed benchmarks, Diff5T serves as a valuable resource for advancing human brain mapping research using diffusion MRI, fostering reproducibility, and enabling collaboration across the neuroscience and medical imaging communities.
Guided MRI Reconstruction via Schrödinger Bridge
Nov 21, 2024



Abstract:Magnetic Resonance Imaging (MRI) is a multi-contrast imaging technique in which different contrast images share similar structural information. However, conventional diffusion models struggle to effectively leverage this structural similarity. Recently, the Schr\"odinger Bridge (SB), a nonlinear extension of the diffusion model, has been proposed to establish diffusion paths between any distributions, allowing the incorporation of guided priors. This study proposes an SB-based, multi-contrast image-guided reconstruction framework that establishes a diffusion bridge between the guiding and target image distributions. By using the guiding image along with data consistency during sampling, the target image is reconstructed more accurately. To better address structural differences between images, we introduce an inversion strategy from the field of image editing, termed $\mathbf{I}^2$SB-inversion. Experiments on a paried T1 and T2-FLAIR datasets demonstrate that $\mathbf{I}^2$SB-inversion achieve a high acceleration up to 14.4 and outperforms existing methods in terms of both reconstruction accuracy and stability.
SamRobNODDI: Q-Space Sampling-Augmented Continuous Representation Learning for Robust and Generalized NODDI
Nov 10, 2024Abstract:Neurite Orientation Dispersion and Density Imaging (NODDI) microstructure estimation from diffusion magnetic resonance imaging (dMRI) is of great significance for the discovery and treatment of various neurological diseases. Current deep learning-based methods accelerate the speed of NODDI parameter estimation and improve the accuracy. However, most methods require the number and coordinates of gradient directions during testing and training to remain strictly consistent, significantly limiting the generalization and robustness of these models in NODDI parameter estimation. In this paper, we propose a q-space sampling augmentation-based continuous representation learning framework (SamRobNODDI) to achieve robust and generalized NODDI. Specifically, a continuous representation learning method based on q-space sampling augmentation is introduced to fully explore the information between different gradient directions in q-space. Furthermore, we design a sampling consistency loss to constrain the outputs of different sampling schemes, ensuring that the outputs remain as consistent as possible, thereby further enhancing performance and robustness to varying q-space sampling schemes. SamRobNODDI is also a flexible framework that can be applied to different backbone networks. To validate the effectiveness of the proposed method, we compared it with 7 state-of-the-art methods across 18 different q-space sampling schemes, demonstrating that the proposed SamRobNODDI has better performance, robustness, generalization, and flexibility.
Quantum Neural Network for Accelerated Magnetic Resonance Imaging
Oct 12, 2024



Abstract:Magnetic resonance image reconstruction starting from undersampled k-space data requires the recovery of many potential nonlinear features, which is very difficult for algorithms to recover these features. In recent years, the development of quantum computing has discovered that quantum convolution can improve network accuracy, possibly due to potential quantum advantages. This article proposes a hybrid neural network containing quantum and classical networks for fast magnetic resonance imaging, and conducts experiments on a quantum computer simulation system. The experimental results indicate that the hybrid network has achieved excellent reconstruction results, and also confirm the feasibility of applying hybrid quantum-classical neural networks into the image reconstruction of rapid magnetic resonance imaging.
Diff-DTI: Fast Diffusion Tensor Imaging Using A Feature-Enhanced Joint Diffusion Model
May 24, 2024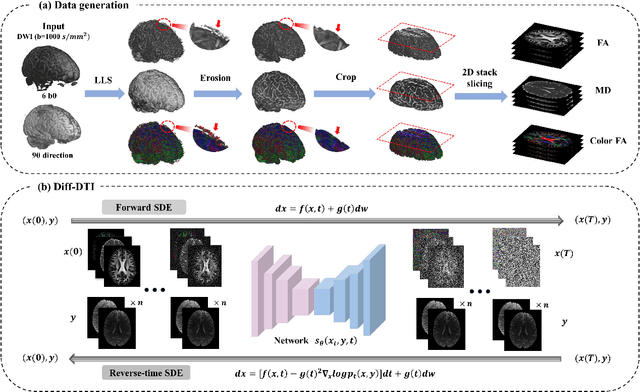
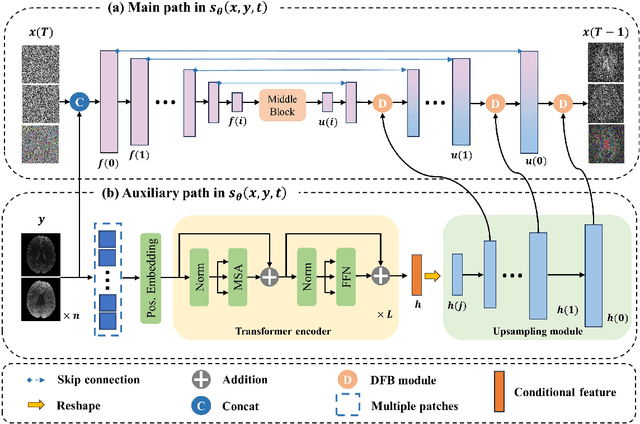
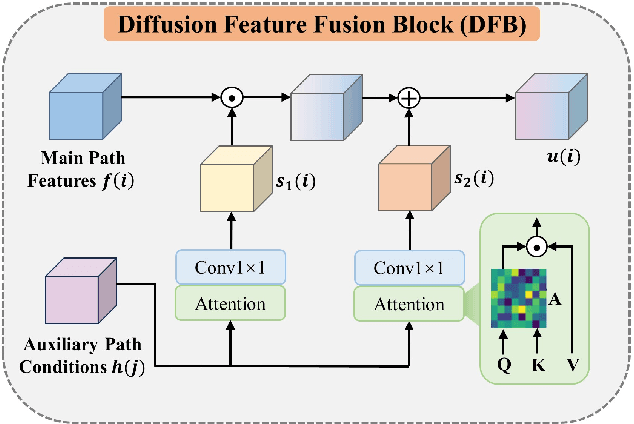
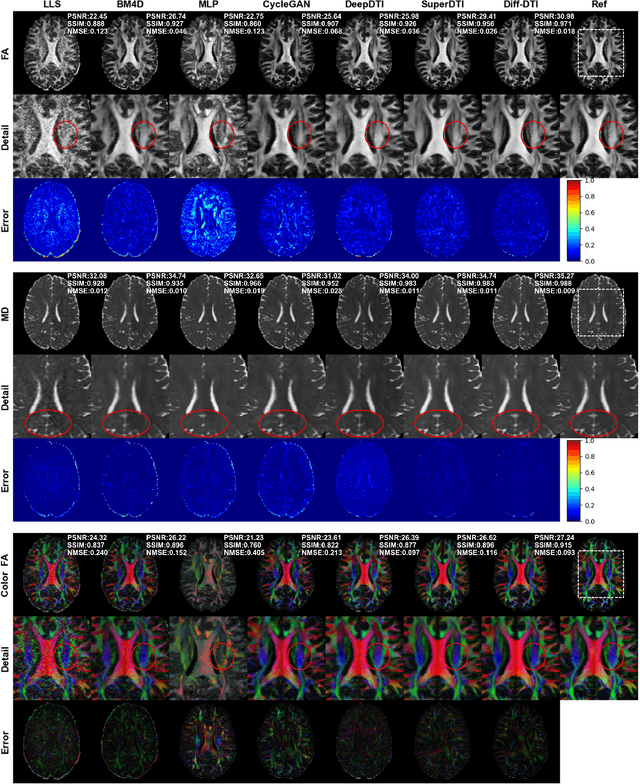
Abstract:Magnetic resonance diffusion tensor imaging (DTI) is a critical tool for neural disease diagnosis. However, long scan time greatly hinders the widespread clinical use of DTI. To accelerate image acquisition, a feature-enhanced joint diffusion model (Diff-DTI) is proposed to obtain accurate DTI parameter maps from a limited number of diffusion-weighted images (DWIs). Diff-DTI introduces a joint diffusion model that directly learns the joint probability distribution of DWIs with DTI parametric maps for conditional generation. Additionally, a feature enhancement fusion mechanism (FEFM) is designed and incorporated into the generative process of Diff-DTI to preserve fine structures in the generated DTI maps. A comprehensive evaluation of the performance of Diff-DTI was conducted on the Human Connectome Project dataset. The results demonstrate that Diff-DTI outperforms existing state-of-the-art fast DTI imaging methods in terms of visual quality and quantitative metrics. Furthermore, Diff-DTI has shown the ability to produce high-fidelity DTI maps with only three DWIs, thus overcoming the requirement of a minimum of six DWIs for DTI.
DeepMpMRI: Tensor-decomposition Regularized Learning for Fast and High-Fidelity Multi-Parametric Microstructural MR Imaging
May 06, 2024Abstract:Deep learning has emerged as a promising approach for learning the nonlinear mapping between diffusion-weighted MR images and tissue parameters, which enables automatic and deep understanding of the brain microstructures. However, the efficiency and accuracy in the multi-parametric estimations are still limited since previous studies tend to estimate multi-parametric maps with dense sampling and isolated signal modeling. This paper proposes DeepMpMRI, a unified framework for fast and high-fidelity multi-parametric estimation from various diffusion models using sparsely sampled q-space data. DeepMpMRI is equipped with a newly designed tensor-decomposition-based regularizer to effectively capture fine details by exploiting the correlation across parameters. In addition, we introduce a Nesterov-based adaptive learning algorithm that optimizes the regularization parameter dynamically to enhance the performance. DeepMpMRI is an extendable framework capable of incorporating flexible network architecture. Experimental results demonstrate the superiority of our approach over 5 state-of-the-art methods in simultaneously estimating multi-parametric maps for various diffusion models with fine-grained details both quantitatively and qualitatively, achieving 4.5 - 22.5$\times$ acceleration compared to the dense sampling of a total of 270 diffusion gradients.
Pathology-genomic fusion via biologically informed cross-modality graph learning for survival analysis
Apr 11, 2024



Abstract:The diagnosis and prognosis of cancer are typically based on multi-modal clinical data, including histology images and genomic data, due to the complex pathogenesis and high heterogeneity. Despite the advancements in digital pathology and high-throughput genome sequencing, establishing effective multi-modal fusion models for survival prediction and revealing the potential association between histopathology and transcriptomics remains challenging. In this paper, we propose Pathology-Genome Heterogeneous Graph (PGHG) that integrates whole slide images (WSI) and bulk RNA-Seq expression data with heterogeneous graph neural network for cancer survival analysis. The PGHG consists of biological knowledge-guided representation learning network and pathology-genome heterogeneous graph. The representation learning network utilizes the biological prior knowledge of intra-modal and inter-modal data associations to guide the feature extraction. The node features of each modality are updated through attention-based graph learning strategy. Unimodal features and bi-modal fused features are extracted via attention pooling module and then used for survival prediction. We evaluate the model on low-grade gliomas, glioblastoma, and kidney renal papillary cell carcinoma datasets from the Cancer Genome Atlas (TCGA) and the First Affiliated Hospital of Zhengzhou University (FAHZU). Extensive experimental results demonstrate that the proposed method outperforms both unimodal and other multi-modal fusion models. For demonstrating the model interpretability, we also visualize the attention heatmap of pathological images and utilize integrated gradient algorithm to identify important tissue structure, biological pathways and key genes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge