Wenxin Fan
3D Anatomical Structure-guided Deep Learning for Accurate Diffusion Microstructure Imaging
Feb 25, 2025Abstract:Diffusion magnetic resonance imaging (dMRI) is a crucial non-invasive technique for exploring the microstructure of the living human brain. Traditional hand-crafted and model-based tissue microstructure reconstruction methods often require extensive diffusion gradient sampling, which can be time-consuming and limits the clinical applicability of tissue microstructure information. Recent advances in deep learning have shown promise in microstructure estimation; however, accurately estimating tissue microstructure from clinically feasible dMRI scans remains challenging without appropriate constraints. This paper introduces a novel framework that achieves high-fidelity and rapid diffusion microstructure imaging by simultaneously leveraging anatomical information from macro-level priors and mutual information across parameters. This approach enhances time efficiency while maintaining accuracy in microstructure estimation. Experimental results demonstrate that our method outperforms four state-of-the-art techniques, achieving a peak signal-to-noise ratio (PSNR) of 30.51$\pm$0.58 and a structural similarity index measure (SSIM) of 0.97$\pm$0.004 in estimating parametric maps of multiple diffusion models. Notably, our method achieves a 15$\times$ acceleration compared to the dense sampling approach, which typically utilizes 270 diffusion gradients.
Spatial-Angular Representation Learning for High-Fidelity Continuous Super-Resolution in Diffusion MRI
Jan 27, 2025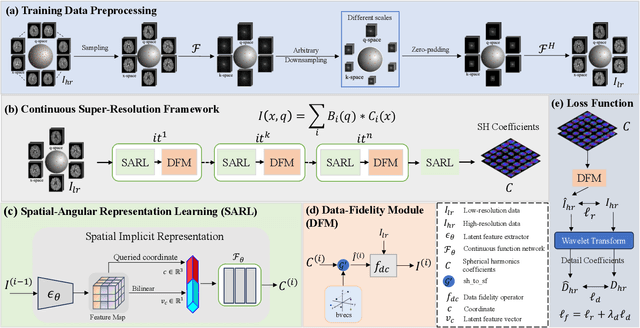
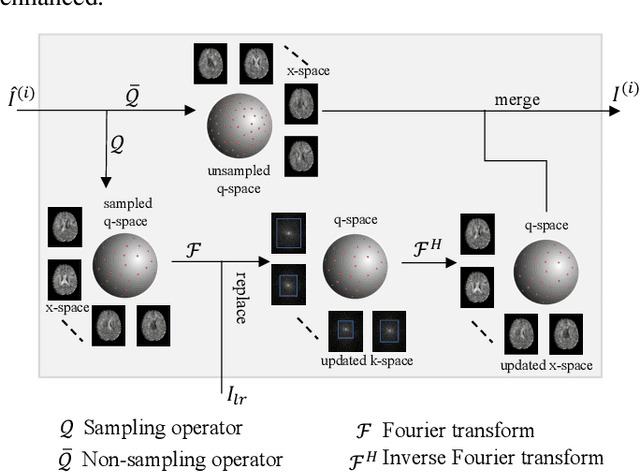
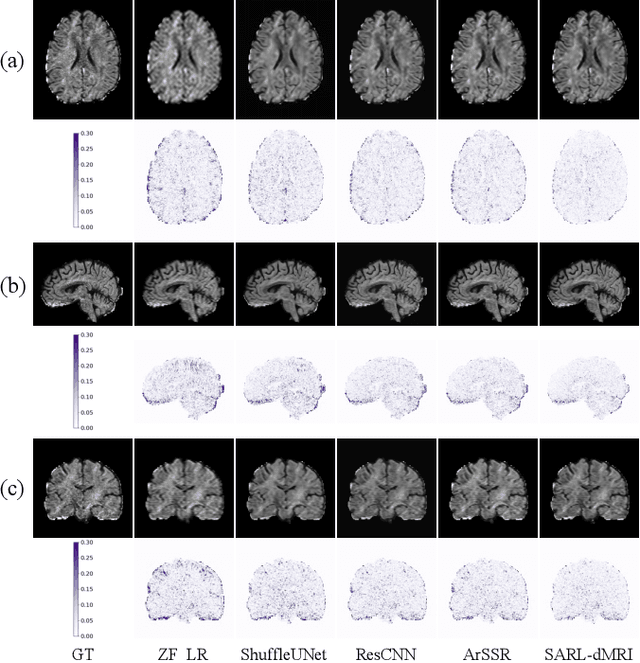
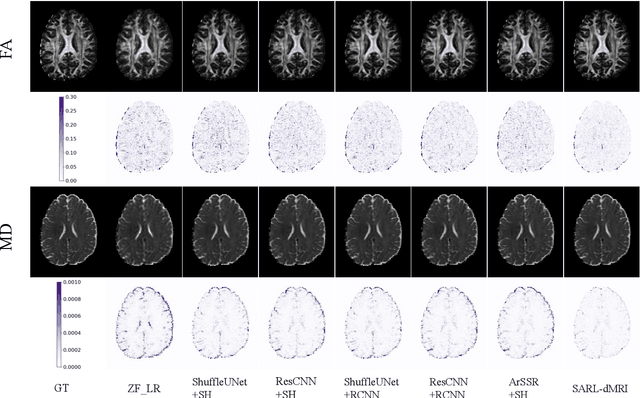
Abstract:Diffusion magnetic resonance imaging (dMRI) often suffers from low spatial and angular resolution due to inherent limitations in imaging hardware and system noise, adversely affecting the accurate estimation of microstructural parameters with fine anatomical details. Deep learning-based super-resolution techniques have shown promise in enhancing dMRI resolution without increasing acquisition time. However, most existing methods are confined to either spatial or angular super-resolution, limiting their effectiveness in capturing detailed microstructural features. Furthermore, traditional pixel-wise loss functions struggle to recover intricate image details essential for high-resolution reconstruction. To address these challenges, we propose SARL-dMRI, a novel Spatial-Angular Representation Learning framework for high-fidelity, continuous super-resolution in dMRI. SARL-dMRI explores implicit neural representations and spherical harmonics to model continuous spatial and angular representations, simultaneously enhancing both spatial and angular resolution while improving microstructural parameter estimation accuracy. To further preserve image fidelity, a data-fidelity module and wavelet-based frequency loss are introduced, ensuring the super-resolved images remain consistent with the original input and retain fine details. Extensive experiments demonstrate that, compared to five other state-of-the-art methods, our method significantly enhances dMRI data resolution, improves the accuracy of microstructural parameter estimation, and provides better generalization capabilities. It maintains stable performance even under a 45$\times$ downsampling factor.
SamRobNODDI: Q-Space Sampling-Augmented Continuous Representation Learning for Robust and Generalized NODDI
Nov 10, 2024Abstract:Neurite Orientation Dispersion and Density Imaging (NODDI) microstructure estimation from diffusion magnetic resonance imaging (dMRI) is of great significance for the discovery and treatment of various neurological diseases. Current deep learning-based methods accelerate the speed of NODDI parameter estimation and improve the accuracy. However, most methods require the number and coordinates of gradient directions during testing and training to remain strictly consistent, significantly limiting the generalization and robustness of these models in NODDI parameter estimation. In this paper, we propose a q-space sampling augmentation-based continuous representation learning framework (SamRobNODDI) to achieve robust and generalized NODDI. Specifically, a continuous representation learning method based on q-space sampling augmentation is introduced to fully explore the information between different gradient directions in q-space. Furthermore, we design a sampling consistency loss to constrain the outputs of different sampling schemes, ensuring that the outputs remain as consistent as possible, thereby further enhancing performance and robustness to varying q-space sampling schemes. SamRobNODDI is also a flexible framework that can be applied to different backbone networks. To validate the effectiveness of the proposed method, we compared it with 7 state-of-the-art methods across 18 different q-space sampling schemes, demonstrating that the proposed SamRobNODDI has better performance, robustness, generalization, and flexibility.
RobNODDI: Robust NODDI Parameter Estimation with Adaptive Sampling under Continuous Representation
Aug 04, 2024



Abstract:Neurite Orientation Dispersion and Density Imaging (NODDI) is an important imaging technology used to evaluate the microstructure of brain tissue, which is of great significance for the discovery and treatment of various neurological diseases. Current deep learning-based methods perform parameter estimation through diffusion magnetic resonance imaging (dMRI) with a small number of diffusion gradients. These methods speed up parameter estimation and improve accuracy. However, the diffusion directions used by most existing deep learning models during testing needs to be strictly consistent with the diffusion directions during training. This results in poor generalization and robustness of deep learning models in dMRI parameter estimation. In this work, we verify for the first time that the parameter estimation performance of current mainstream methods will significantly decrease when the testing diffusion directions and the training diffusion directions are inconsistent. A robust NODDI parameter estimation method with adaptive sampling under continuous representation (RobNODDI) is proposed. Furthermore, long short-term memory (LSTM) units and fully connected layers are selected to learn continuous representation signals. To this end, we use a total of 100 subjects to conduct experiments based on the Human Connectome Project (HCP) dataset, of which 60 are used for training, 20 are used for validation, and 20 are used for testing. The test results indicate that RobNODDI improves the generalization performance and robustness of the deep learning model, enhancing the stability and flexibility of deep learning NODDI parameter estimatimation applications.
DeepMpMRI: Tensor-decomposition Regularized Learning for Fast and High-Fidelity Multi-Parametric Microstructural MR Imaging
May 06, 2024Abstract:Deep learning has emerged as a promising approach for learning the nonlinear mapping between diffusion-weighted MR images and tissue parameters, which enables automatic and deep understanding of the brain microstructures. However, the efficiency and accuracy in the multi-parametric estimations are still limited since previous studies tend to estimate multi-parametric maps with dense sampling and isolated signal modeling. This paper proposes DeepMpMRI, a unified framework for fast and high-fidelity multi-parametric estimation from various diffusion models using sparsely sampled q-space data. DeepMpMRI is equipped with a newly designed tensor-decomposition-based regularizer to effectively capture fine details by exploiting the correlation across parameters. In addition, we introduce a Nesterov-based adaptive learning algorithm that optimizes the regularization parameter dynamically to enhance the performance. DeepMpMRI is an extendable framework capable of incorporating flexible network architecture. Experimental results demonstrate the superiority of our approach over 5 state-of-the-art methods in simultaneously estimating multi-parametric maps for various diffusion models with fine-grained details both quantitatively and qualitatively, achieving 4.5 - 22.5$\times$ acceleration compared to the dense sampling of a total of 270 diffusion gradients.
CSR-dMRI: Continuous Super-Resolution of Diffusion MRI with Anatomical Structure-assisted Implicit Neural Representation Learning
Apr 04, 2024Abstract:Deep learning-based dMRI super-resolution methods can effectively enhance image resolution by leveraging the learning capabilities of neural networks on large datasets. However, these methods tend to learn a fixed scale mapping between low-resolution (LR) and high-resolution (HR) images, overlooking the need for radiologists to scale the images at arbitrary resolutions. Moreover, the pixel-wise loss in the image domain tends to generate over-smoothed results, losing fine textures and edge information. To address these issues, we propose a novel continuous super-resolution of dMRI with anatomical structure-assisted implicit neural representation learning method, called CSR-dMRI. Specifically, the CSR-dMRI model consists of two components. The first is the latent feature extractor, which primarily extracts latent space feature maps from LR dMRI and anatomical images while learning structural prior information from the anatomical images. The second is the implicit function network, which utilizes voxel coordinates and latent feature vectors to generate voxel intensities at corresponding positions. Additionally, a frequency-domain-based loss is introduced to preserve the structural and texture information, further enhancing the image quality. Extensive experiments on the publicly available HCP dataset validate the effectiveness of our approach. Furthermore, our method demonstrates superior generalization capability and can be applied to arbitrary-scale super-resolution, including non-integer scale factors, expanding its applicability beyond conventional approaches.
Sequential Model for Predicting Patient Adherence in Subcutaneous Immunotherapy for Allergic Rhinitis
Jan 23, 2024Abstract:Objective: Subcutaneous Immunotherapy (SCIT) is the long-lasting causal treatment of allergic rhinitis. How to enhance the adherence of patients to maximize the benefit of allergen immunotherapy (AIT) plays a crucial role in the management of AIT. This study aims to leverage novel machine learning models to precisely predict the risk of non-adherence of patients and related systematic symptom scores, to provide a novel approach in the management of long-term AIT. Methods: The research develops and analyzes two models, Sequential Latent Actor-Critic (SLAC) and Long Short-Term Memory (LSTM), evaluating them based on scoring and adherence prediction capabilities. Results: Excluding the biased samples at the first time step, the predictive adherence accuracy of the SLAC models is from $60\,\%$ to $72\%$, and for LSTM models, it is $66\,\%$ to $84\,\%$, varying according to the time steps. The range of Root Mean Square Error (RMSE) for SLAC models is between $0.93$ and $2.22$, while for LSTM models it is between $1.09$ and $1.77$. Notably, these RMSEs are significantly lower than the random prediction error of $4.55$. Conclusion: We creatively apply sequential models in the long-term management of SCIT with promising accuracy in the prediction of SCIT nonadherence in Allergic Rhinitis (AR) patients. While LSTM outperforms SLAC in adherence prediction, SLAC excels in score prediction for patients undergoing SCIT for AR. The state-action-based SLAC adds flexibility, presenting a novel and effective approach for managing long-term AIT.
AID-DTI: Accelerating High-fidelity Diffusion Tensor Imaging with Detail-Preserving Model-based Deep Learning
Jan 03, 2024Abstract:Deep learning has shown great potential in accelerating diffusion tensor imaging (DTI). Nevertheless, existing methods tend to suffer from Rician noise and detail loss in reconstructing the DTI-derived parametric maps especially when sparsely sampled q-space data are used. This paper proposes a novel method, AID-DTI (Accelerating hIgh fiDelity Diffusion Tensor Imaging), to facilitate fast and accurate DTI with only six measurements. AID-DTI is equipped with a newly designed Singular Value Decomposition (SVD)-based regularizer, which can effectively capture fine details while suppressing noise during network training. Experimental results on Human Connectome Project (HCP) data consistently demonstrate that the proposed method estimates DTI parameter maps with fine-grained details and outperforms three state-of-the-art methods both quantitatively and qualitatively.
Simultaneous q-Space Sampling Optimization and Reconstruction for Fast and High-fidelity Diffusion Magnetic Resonance Imaging
Jan 03, 2024Abstract:Diffusion Magnetic Resonance Imaging (dMRI) plays a crucial role in the noninvasive investigation of tissue microstructural properties and structural connectivity in the \textit{in vivo} human brain. However, to effectively capture the intricate characteristics of water diffusion at various directions and scales, it is important to employ comprehensive q-space sampling. Unfortunately, this requirement leads to long scan times, limiting the clinical applicability of dMRI. To address this challenge, we propose SSOR, a Simultaneous q-Space sampling Optimization and Reconstruction framework. We jointly optimize a subset of q-space samples using a continuous representation of spherical harmonic functions and a reconstruction network. Additionally, we integrate the unique properties of diffusion magnetic resonance imaging (dMRI) in both the q-space and image domains by applying $l1$-norm and total-variation regularization. The experiments conducted on HCP data demonstrate that SSOR has promising strengths both quantitatively and qualitatively and exhibits robustness to noise.
Rethinking the optimization process for self-supervised model-driven MRI reconstruction
Mar 18, 2022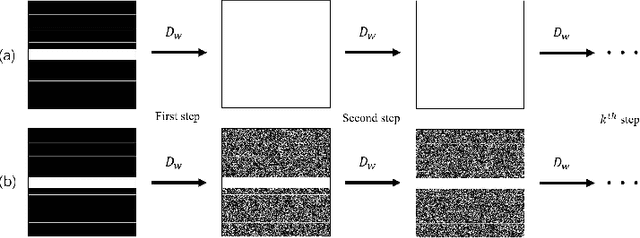
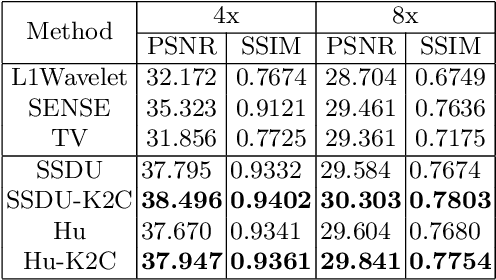
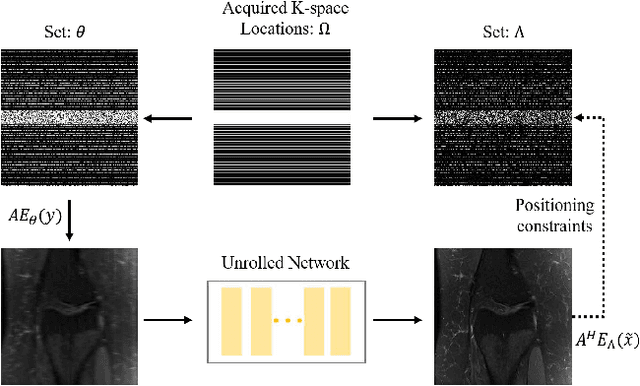

Abstract:Recovering high-quality images from undersampled measurements is critical for accelerated MRI reconstruction. Recently, various supervised deep learning-based MRI reconstruction methods have been developed. Despite the achieved promising performances, these methods require fully sampled reference data, the acquisition of which is resource-intensive and time-consuming. Self-supervised learning has emerged as a promising solution to alleviate the reliance on fully sampled datasets. However, existing self-supervised methods suffer from reconstruction errors due to the insufficient constraint enforced on the non-sampled data points and the error accumulation happened alongside the iterative image reconstruction process for model-driven deep learning reconstrutions. To address these challenges, we propose K2Calibrate, a K-space adaptation strategy for self-supervised model-driven MR reconstruction optimization. By iteratively calibrating the learned measurements, K2Calibrate can reduce the network's reconstruction deterioration caused by statistically dependent noise. Extensive experiments have been conducted on the open-source dataset FastMRI, and K2Calibrate achieves better results than five state-of-the-art methods. The proposed K2Calibrate is plug-and-play and can be easily integrated with different model-driven deep learning reconstruction methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge