Taohui Xiao
SamRobNODDI: Q-Space Sampling-Augmented Continuous Representation Learning for Robust and Generalized NODDI
Nov 10, 2024Abstract:Neurite Orientation Dispersion and Density Imaging (NODDI) microstructure estimation from diffusion magnetic resonance imaging (dMRI) is of great significance for the discovery and treatment of various neurological diseases. Current deep learning-based methods accelerate the speed of NODDI parameter estimation and improve the accuracy. However, most methods require the number and coordinates of gradient directions during testing and training to remain strictly consistent, significantly limiting the generalization and robustness of these models in NODDI parameter estimation. In this paper, we propose a q-space sampling augmentation-based continuous representation learning framework (SamRobNODDI) to achieve robust and generalized NODDI. Specifically, a continuous representation learning method based on q-space sampling augmentation is introduced to fully explore the information between different gradient directions in q-space. Furthermore, we design a sampling consistency loss to constrain the outputs of different sampling schemes, ensuring that the outputs remain as consistent as possible, thereby further enhancing performance and robustness to varying q-space sampling schemes. SamRobNODDI is also a flexible framework that can be applied to different backbone networks. To validate the effectiveness of the proposed method, we compared it with 7 state-of-the-art methods across 18 different q-space sampling schemes, demonstrating that the proposed SamRobNODDI has better performance, robustness, generalization, and flexibility.
RobNODDI: Robust NODDI Parameter Estimation with Adaptive Sampling under Continuous Representation
Aug 04, 2024



Abstract:Neurite Orientation Dispersion and Density Imaging (NODDI) is an important imaging technology used to evaluate the microstructure of brain tissue, which is of great significance for the discovery and treatment of various neurological diseases. Current deep learning-based methods perform parameter estimation through diffusion magnetic resonance imaging (dMRI) with a small number of diffusion gradients. These methods speed up parameter estimation and improve accuracy. However, the diffusion directions used by most existing deep learning models during testing needs to be strictly consistent with the diffusion directions during training. This results in poor generalization and robustness of deep learning models in dMRI parameter estimation. In this work, we verify for the first time that the parameter estimation performance of current mainstream methods will significantly decrease when the testing diffusion directions and the training diffusion directions are inconsistent. A robust NODDI parameter estimation method with adaptive sampling under continuous representation (RobNODDI) is proposed. Furthermore, long short-term memory (LSTM) units and fully connected layers are selected to learn continuous representation signals. To this end, we use a total of 100 subjects to conduct experiments based on the Human Connectome Project (HCP) dataset, of which 60 are used for training, 20 are used for validation, and 20 are used for testing. The test results indicate that RobNODDI improves the generalization performance and robustness of the deep learning model, enhancing the stability and flexibility of deep learning NODDI parameter estimatimation applications.
Parameter Constrained Transfer Learning for Low Dose PET Image Denoising
Oct 13, 2019



Abstract:Positron emission tomography (PET) is widely used in clinical practice. However, the potential risk of PET-associated radiation dose to patients needs to be minimized. With reduction of the radiation dose, the resultant images may suffer from noise and artifacts which compromises the diagnostic performance. In this paper, we propose a parameter-constrained generative adversarial network with Wasserstein distance and perceptual loss (PC-WGAN) for low-dose PET image denoising. This method makes two main contributions: 1) a PC-WGAN framework is designed to denoise low-dose PET images without compromising structural details; and 2) a transfer learning strategy is developed to train PC-WGAN with parameters being constrained, which has major merits; namely, making the training process of PC-WGAN efficient and improving the quality of denoised images. The experimental results on clinical data show that the proposed network can suppress image noise more effectively while preserving better image fidelity than three selected state-of-the-art methods.
LANTERN: learn analysis transform network for dynamic magnetic resonance imaging with small dataset
Aug 24, 2019



Abstract:This paper proposes to learn analysis transform network for dynamic magnetic resonance imaging (LANTERN) with small dataset. Integrating the strength of CS-MRI and deep learning, the proposed framework is highlighted in three components: (i) The spatial and temporal domains are sparsely constrained by using adaptively trained CNN. (ii) We introduce an end-to-end framework to learn the parameters in LANTERN to solve the difficulty of parameter selection in traditional methods. (iii) Compared to existing deep learning reconstruction methods, our reconstruction accuracy is better when the amount of data is limited. Our model is able to fully exploit the redundancy in spatial and temporal of dynamic MR images. We performed quantitative and qualitative analysis of cardiac datasets at different acceleration factors (2x-11x) and different undersampling modes. In comparison with state-of-the-art methods, extensive experiments show that our method achieves consistent better reconstruction performance on the MRI reconstruction in terms of three quantitative metrics (PSNR, SSIM and HFEN) under different undersamling patterns and acceleration factors.
Model-based Convolutional De-Aliasing Network Learning for Parallel MR Imaging
Aug 06, 2019



Abstract:Parallel imaging has been an essential technique to accelerate MR imaging. Nevertheless, the acceleration rate is still limited due to the ill-condition and challenges associated with the undersampled reconstruction. In this paper, we propose a model-based convolutional de-aliasing network with adaptive parameter learning to achieve accurate reconstruction from multi-coil undersampled k-space data. Three main contributions have been made: a de-aliasing reconstruction model was proposed to accelerate parallel MR imaging with deep learning exploring both spatial redundancy and multi-coil correlations; a split Bregman iteration algorithm was developed to solve the model efficiently; and unlike most existing parallel imaging methods which rely on the accuracy of the estimated multi-coil sensitivity, the proposed method can perform parallel reconstruction from undersampled data without explicit sensitivity calculation. Evaluations were conducted on \emph{in vivo} brain dataset with a variety of undersampling patterns and different acceleration factors. Our results demonstrated that this method could achieve superior performance in both quantitative and qualitative analysis, compared to three state-of-the-art methods.
DeepcomplexMRI: Exploiting deep residual network for fast parallel MR imaging with complex convolution
Jul 29, 2019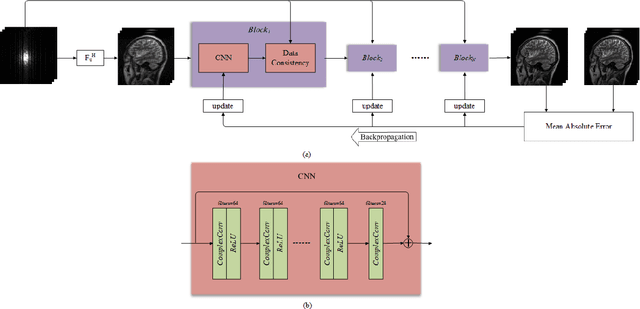
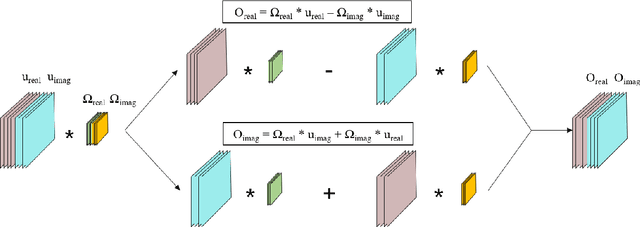

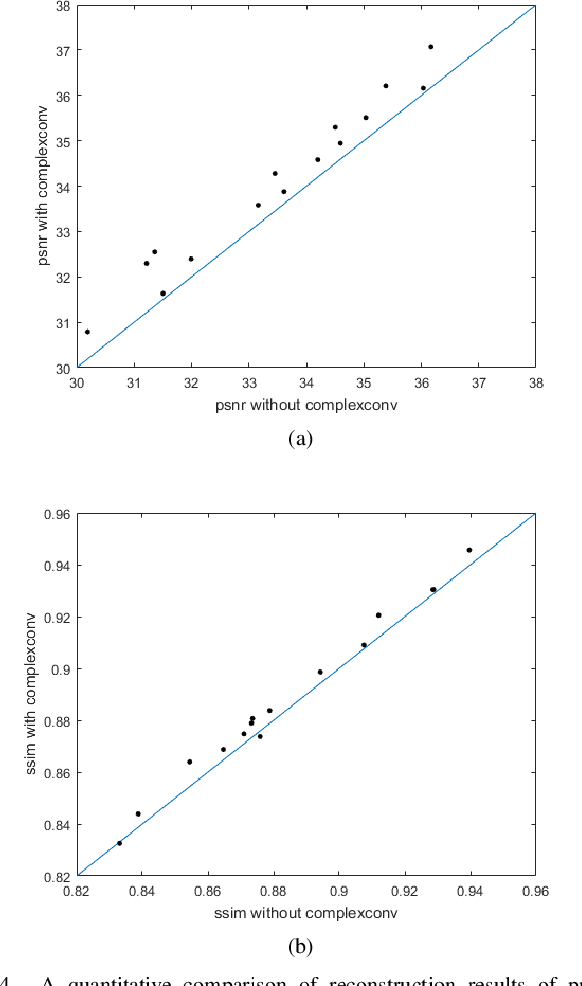
Abstract:This paper proposes a multi-channel image reconstruction method, named DeepcomplexMRI, to accelerate parallel MR imaging with residual complex convolutional neural network. Different from most existing works which rely on the utilization of the coil sensitivities or prior information of predefined transforms, DeepcomplexMRI takes advantage of the availability of a large number of existing multi-channel groudtruth images and uses them as labeled data to train the deep residual convolutional neural network offline. In particular, a complex convolutional network is proposed to take into account the correlation between the real and imaginary parts of MR images. In addition, the k space data consistency is further enforced repeatedly in between layers of the network. The evaluations on in vivo datasets show that the proposed method has the capability to recover the desired multi-channel images. Its comparison with state-of-the-art method also demonstrates that the proposed method can reconstruct the desired MR images more accurately.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge