Dong Liang
Fully Kolmogorov-Arnold Deep Model in Medical Image Segmentation
Feb 03, 2026Abstract:Deeply stacked KANs are practically impossible due to high training difficulties and substantial memory requirements. Consequently, existing studies can only incorporate few KAN layers, hindering the comprehensive exploration of KANs. This study overcomes these limitations and introduces the first fully KA-based deep model, demonstrating that KA-based layers can entirely replace traditional architectures in deep learning and achieve superior learning capacity. Specifically, (1) the proposed Share-activation KAN (SaKAN) reformulates Sprecher's variant of Kolmogorov-Arnold representation theorem, which achieves better optimization due to its simplified parameterization and denser training samples, to ease training difficulty, (2) this paper indicates that spline gradients contribute negligibly to training while consuming huge GPU memory, thus proposes the Grad-Free Spline to significantly reduce memory usage and computational overhead. (3) Building on these two innovations, our ALL U-KAN is the first representative implementation of fully KA-based deep model, where the proposed KA and KAonv layers completely replace FC and Conv layers. Extensive evaluations on three medical image segmentation tasks confirm the superiority of the full KA-based architecture compared to partial KA-based and traditional architectures, achieving all higher segmentation accuracy. Compared to directly deeply stacked KAN, ALL U-KAN achieves 10 times reduction in parameter count and reduces memory consumption by more than 20 times, unlocking the new explorations into deep KAN architectures.
Beyond Dense States: Elevating Sparse Transcoders to Active Operators for Latent Reasoning
Feb 02, 2026Abstract:Latent reasoning compresses the chain-of-thought (CoT) into continuous hidden states, yet existing methods rely on dense latent transitions that remain difficult to interpret and control. Meanwhile, sparse representation models uncover human-interpretable semantic features but remain largely confined to post-hoc analysis. We reconcile this tension by proposing LSTR (Latent Sparse Transcoder Reasoning), a latent reasoning framework that elevates functional sparse transcoders into active reasoning operators to perform multi-step computation through sparse semantic transitions. At its core, LSTR employs a Latent Transition Transcoder (LTT) with a residual skip architecture that decouples linear manifold transport from sparse semantic updates, enabling controllable semantic resolution via explicit sparsity constraints. Extensive experiments show that LSTR preserves reasoning accuracy and compression efficiency while substantially improving interpretability over dense latent baselines. Causal interventions and trajectory analyses further demonstrate that these sparse features act as both interpretable and causally effective operators in the reasoning process.
World-Shaper: A Unified Framework for 360° Panoramic Editing
Jan 30, 2026Abstract:Being able to edit panoramic images is crucial for creating realistic 360° visual experiences. However, existing perspective-based image editing methods fail to model the spatial structure of panoramas. Conventional cube-map decompositions attempt to overcome this problem but inevitably break global consistency due to their mismatch with spherical geometry. Motivated by this insight, we reformulate panoramic editing directly in the equirectangular projection (ERP) domain and present World-Shaper, a unified geometry-aware framework that bridges panoramic generation and editing within a single editing-centric design. To overcome the scarcity of paired data, we adopt a generate-then-edit paradigm, where controllable panoramic generation serves as an auxiliary stage to synthesize diverse paired examples for supervised editing learning. To address geometric distortion, we introduce a geometry-aware learning strategy that explicitly enforces position-aware shape supervision and implicitly internalizes panoramic priors through progressive training. Extensive experiments on our new benchmark, PEBench, demonstrate that our method achieves superior geometric consistency, editing fidelity, and text controllability compared to SOTA methods, enabling coherent and flexible 360° visual world creation with unified editing control. Code, model, and data will be released at our project page: https://world-shaper-project.github.io/
Meta Lattice: Model Space Redesign for Cost-Effective Industry-Scale Ads Recommendations
Dec 15, 2025Abstract:The rapidly evolving landscape of products, surfaces, policies, and regulations poses significant challenges for deploying state-of-the-art recommendation models at industry scale, primarily due to data fragmentation across domains and escalating infrastructure costs that hinder sustained quality improvements. To address this challenge, we propose Lattice, a recommendation framework centered around model space redesign that extends Multi-Domain, Multi-Objective (MDMO) learning beyond models and learning objectives. Lattice addresses these challenges through a comprehensive model space redesign that combines cross-domain knowledge sharing, data consolidation, model unification, distillation, and system optimizations to achieve significant improvements in both quality and cost-efficiency. Our deployment of Lattice at Meta has resulted in 10% revenue-driving top-line metrics gain, 11.5% user satisfaction improvement, 6% boost in conversion rate, with 20% capacity saving.
RoboOS-NeXT: A Unified Memory-based Framework for Lifelong, Scalable, and Robust Multi-Robot Collaboration
Oct 30, 2025Abstract:The proliferation of collaborative robots across diverse tasks and embodiments presents a central challenge: achieving lifelong adaptability, scalable coordination, and robust scheduling in multi-agent systems. Existing approaches, from vision-language-action (VLA) models to hierarchical frameworks, fall short due to their reliance on limited or dividual-agent memory. This fundamentally constrains their ability to learn over long horizons, scale to heterogeneous teams, or recover from failures, highlighting the need for a unified memory representation. To address these limitations, we introduce RoboOS-NeXT, a unified memory-based framework for lifelong, scalable, and robust multi-robot collaboration. At the core of RoboOS-NeXT is the novel Spatio-Temporal-Embodiment Memory (STEM), which integrates spatial scene geometry, temporal event history, and embodiment profiles into a shared representation. This memory-centric design is integrated into a brain-cerebellum framework, where a high-level brain model performs global planning by retrieving and updating STEM, while low-level controllers execute actions locally. This closed loop between cognition, memory, and execution enables dynamic task allocation, fault-tolerant collaboration, and consistent state synchronization. We conduct extensive experiments spanning complex coordination tasks in restaurants, supermarkets, and households. Our results demonstrate that RoboOS-NeXT achieves superior performance across heterogeneous embodiments, validating its effectiveness in enabling lifelong, scalable, and robust multi-robot collaboration. Project website: https://flagopen.github.io/RoboOS/
NeuroSwift: A Lightweight Cross-Subject Framework for fMRI Visual Reconstruction of Complex Scenes
Oct 02, 2025



Abstract:Reconstructing visual information from brain activity via computer vision technology provides an intuitive understanding of visual neural mechanisms. Despite progress in decoding fMRI data with generative models, achieving accurate cross-subject reconstruction of visual stimuli remains challenging and computationally demanding. This difficulty arises from inter-subject variability in neural representations and the brain's abstract encoding of core semantic features in complex visual inputs. To address these challenges, we propose NeuroSwift, which integrates complementary adapters via diffusion: AutoKL for low-level features and CLIP for semantics. NeuroSwift's CLIP Adapter is trained on Stable Diffusion generated images paired with COCO captions to emulate higher visual cortex encoding. For cross-subject generalization, we pretrain on one subject and then fine-tune only 17 percent of parameters (fully connected layers) for new subjects, while freezing other components. This enables state-of-the-art performance with only one hour of training per subject on lightweight GPUs (three RTX 4090), and it outperforms existing methods.
MultiMedEdit: A Scenario-Aware Benchmark for Evaluating Knowledge Editing in Medical VQA
Aug 09, 2025Abstract:Knowledge editing (KE) provides a scalable approach for updating factual knowledge in large language models without full retraining. While previous studies have demonstrated effectiveness in general domains and medical QA tasks, little attention has been paid to KE in multimodal medical scenarios. Unlike text-only settings, medical KE demands integrating updated knowledge with visual reasoning to support safe and interpretable clinical decisions. To address this gap, we propose MultiMedEdit, the first benchmark tailored to evaluating KE in clinical multimodal tasks. Our framework spans both understanding and reasoning task types, defines a three-dimensional metric suite (reliability, generality, and locality), and supports cross-paradigm comparisons across general and domain-specific models. We conduct extensive experiments under single-editing and lifelong-editing settings. Results suggest that current methods struggle with generalization and long-tail reasoning, particularly in complex clinical workflows. We further present an efficiency analysis (e.g., edit latency, memory footprint), revealing practical trade-offs in real-world deployment across KE paradigms. Overall, MultiMedEdit not only reveals the limitations of current approaches but also provides a solid foundation for developing clinically robust knowledge editing techniques in the future.
Towards Globally Predictable k-Space Interpolation: A White-box Transformer Approach
Aug 06, 2025Abstract:Interpolating missing data in k-space is essential for accelerating imaging. However, existing methods, including convolutional neural network-based deep learning, primarily exploit local predictability while overlooking the inherent global dependencies in k-space. Recently, Transformers have demonstrated remarkable success in natural language processing and image analysis due to their ability to capture long-range dependencies. This inspires the use of Transformers for k-space interpolation to better exploit its global structure. However, their lack of interpretability raises concerns regarding the reliability of interpolated data. To address this limitation, we propose GPI-WT, a white-box Transformer framework based on Globally Predictable Interpolation (GPI) for k-space. Specifically, we formulate GPI from the perspective of annihilation as a novel k-space structured low-rank (SLR) model. The global annihilation filters in the SLR model are treated as learnable parameters, and the subgradients of the SLR model naturally induce a learnable attention mechanism. By unfolding the subgradient-based optimization algorithm of SLR into a cascaded network, we construct the first white-box Transformer specifically designed for accelerated MRI. Experimental results demonstrate that the proposed method significantly outperforms state-of-the-art approaches in k-space interpolation accuracy while providing superior interpretability.
HOComp: Interaction-Aware Human-Object Composition
Jul 22, 2025Abstract:While existing image-guided composition methods may help insert a foreground object onto a user-specified region of a background image, achieving natural blending inside the region with the rest of the image unchanged, we observe that these existing methods often struggle in synthesizing seamless interaction-aware compositions when the task involves human-object interactions. In this paper, we first propose HOComp, a novel approach for compositing a foreground object onto a human-centric background image, while ensuring harmonious interactions between the foreground object and the background person and their consistent appearances. Our approach includes two key designs: (1) MLLMs-driven Region-based Pose Guidance (MRPG), which utilizes MLLMs to identify the interaction region as well as the interaction type (e.g., holding and lefting) to provide coarse-to-fine constraints to the generated pose for the interaction while incorporating human pose landmarks to track action variations and enforcing fine-grained pose constraints; and (2) Detail-Consistent Appearance Preservation (DCAP), which unifies a shape-aware attention modulation mechanism, a multi-view appearance loss, and a background consistency loss to ensure consistent shapes/textures of the foreground and faithful reproduction of the background human. We then propose the first dataset, named Interaction-aware Human-Object Composition (IHOC), for the task. Experimental results on our dataset show that HOComp effectively generates harmonious human-object interactions with consistent appearances, and outperforms relevant methods qualitatively and quantitatively.
Structure and Smoothness Constrained Dual Networks for MR Bias Field Correction
Jul 02, 2025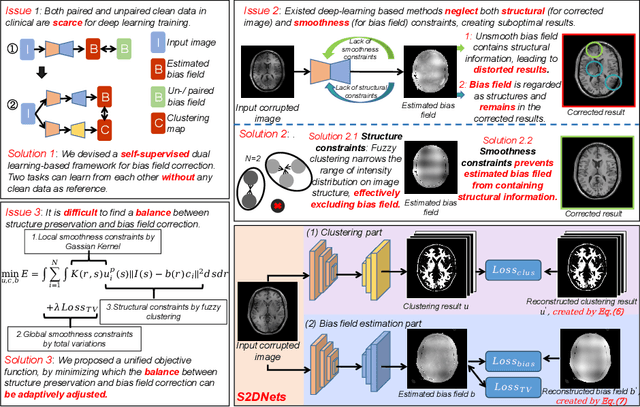
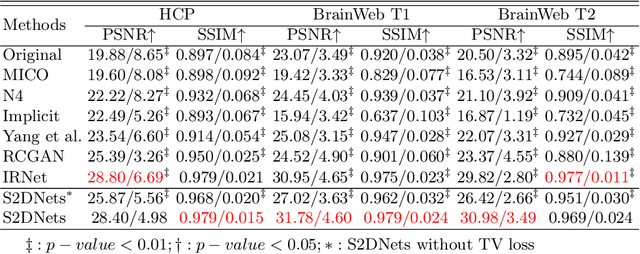
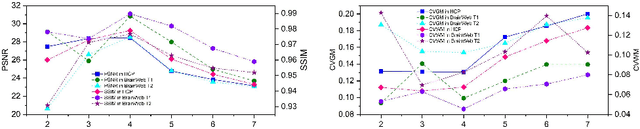
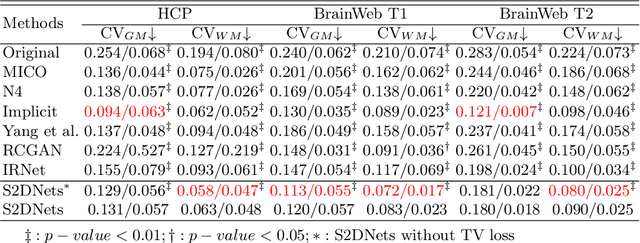
Abstract:MR imaging techniques are of great benefit to disease diagnosis. However, due to the limitation of MR devices, significant intensity inhomogeneity often exists in imaging results, which impedes both qualitative and quantitative medical analysis. Recently, several unsupervised deep learning-based models have been proposed for MR image improvement. However, these models merely concentrate on global appearance learning, and neglect constraints from image structures and smoothness of bias field, leading to distorted corrected results. In this paper, novel structure and smoothness constrained dual networks, named S2DNets, are proposed aiming to self-supervised bias field correction. S2DNets introduce piece-wise structural constraints and smoothness of bias field for network training to effectively remove non-uniform intensity and retain much more structural details. Extensive experiments executed on both clinical and simulated MR datasets show that the proposed model outperforms other conventional and deep learning-based models. In addition to comparison on visual metrics, downstream MR image segmentation tasks are also used to evaluate the impact of the proposed model. The source code is available at: https://github.com/LeongDong/S2DNets}{https://github.com/LeongDong/S2DNets.
* 11 pages, 3 figures, accepted by MICCAI
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge