Kuanquan Wang
Faculty of Computing, Harbin Institute of Technology, Harbin, China
Fully Kolmogorov-Arnold Deep Model in Medical Image Segmentation
Feb 03, 2026Abstract:Deeply stacked KANs are practically impossible due to high training difficulties and substantial memory requirements. Consequently, existing studies can only incorporate few KAN layers, hindering the comprehensive exploration of KANs. This study overcomes these limitations and introduces the first fully KA-based deep model, demonstrating that KA-based layers can entirely replace traditional architectures in deep learning and achieve superior learning capacity. Specifically, (1) the proposed Share-activation KAN (SaKAN) reformulates Sprecher's variant of Kolmogorov-Arnold representation theorem, which achieves better optimization due to its simplified parameterization and denser training samples, to ease training difficulty, (2) this paper indicates that spline gradients contribute negligibly to training while consuming huge GPU memory, thus proposes the Grad-Free Spline to significantly reduce memory usage and computational overhead. (3) Building on these two innovations, our ALL U-KAN is the first representative implementation of fully KA-based deep model, where the proposed KA and KAonv layers completely replace FC and Conv layers. Extensive evaluations on three medical image segmentation tasks confirm the superiority of the full KA-based architecture compared to partial KA-based and traditional architectures, achieving all higher segmentation accuracy. Compared to directly deeply stacked KAN, ALL U-KAN achieves 10 times reduction in parameter count and reduces memory consumption by more than 20 times, unlocking the new explorations into deep KAN architectures.
Ambiguity-aware Truncated Flow Matching for Ambiguous Medical Image Segmentation
Nov 10, 2025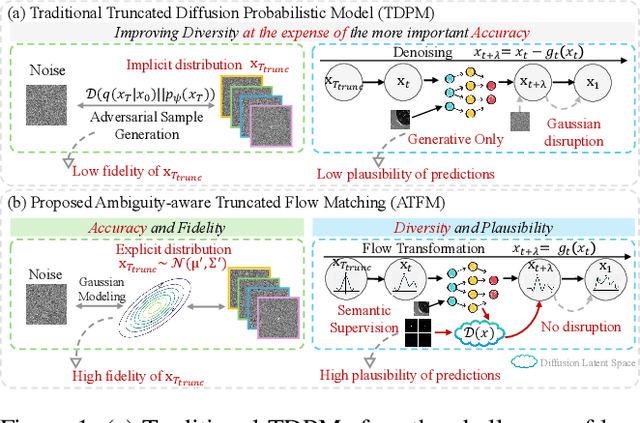
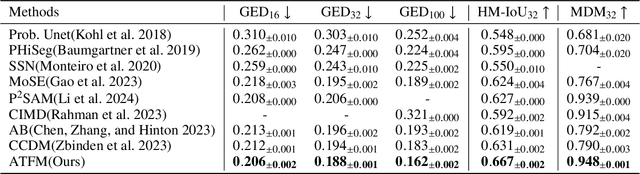
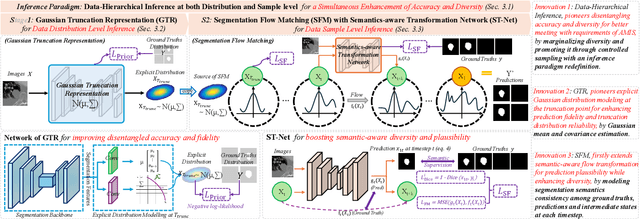
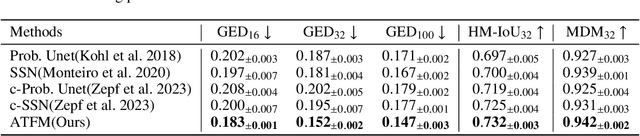
Abstract:A simultaneous enhancement of accuracy and diversity of predictions remains a challenge in ambiguous medical image segmentation (AMIS) due to the inherent trade-offs. While truncated diffusion probabilistic models (TDPMs) hold strong potential with a paradigm optimization, existing TDPMs suffer from entangled accuracy and diversity of predictions with insufficient fidelity and plausibility. To address the aforementioned challenges, we propose Ambiguity-aware Truncated Flow Matching (ATFM), which introduces a novel inference paradigm and dedicated model components. Firstly, we propose Data-Hierarchical Inference, a redefinition of AMIS-specific inference paradigm, which enhances accuracy and diversity at data-distribution and data-sample level, respectively, for an effective disentanglement. Secondly, Gaussian Truncation Representation (GTR) is introduced to enhance both fidelity of predictions and reliability of truncation distribution, by explicitly modeling it as a Gaussian distribution at $T_{\text{trunc}}$ instead of using sampling-based approximations.Thirdly, Segmentation Flow Matching (SFM) is proposed to enhance the plausibility of diverse predictions by extending semantic-aware flow transformation in Flow Matching (FM). Comprehensive evaluations on LIDC and ISIC3 datasets demonstrate that ATFM outperforms SOTA methods and simultaneously achieves a more efficient inference. ATFM improves GED and HM-IoU by up to $12\%$ and $7.3\%$ compared to advanced methods.
Structure and Smoothness Constrained Dual Networks for MR Bias Field Correction
Jul 02, 2025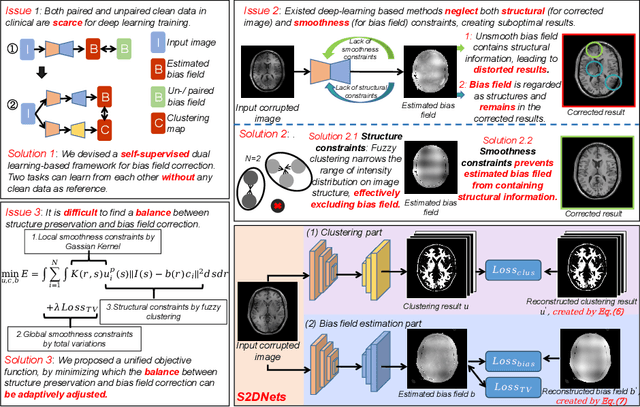
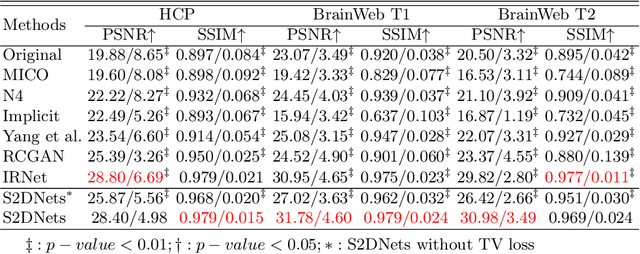
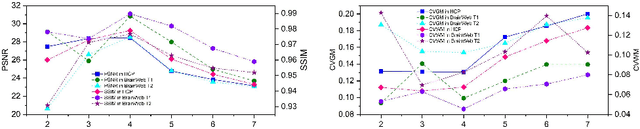
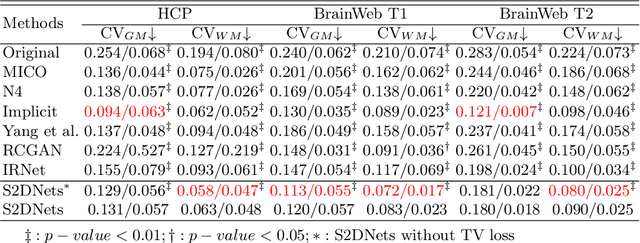
Abstract:MR imaging techniques are of great benefit to disease diagnosis. However, due to the limitation of MR devices, significant intensity inhomogeneity often exists in imaging results, which impedes both qualitative and quantitative medical analysis. Recently, several unsupervised deep learning-based models have been proposed for MR image improvement. However, these models merely concentrate on global appearance learning, and neglect constraints from image structures and smoothness of bias field, leading to distorted corrected results. In this paper, novel structure and smoothness constrained dual networks, named S2DNets, are proposed aiming to self-supervised bias field correction. S2DNets introduce piece-wise structural constraints and smoothness of bias field for network training to effectively remove non-uniform intensity and retain much more structural details. Extensive experiments executed on both clinical and simulated MR datasets show that the proposed model outperforms other conventional and deep learning-based models. In addition to comparison on visual metrics, downstream MR image segmentation tasks are also used to evaluate the impact of the proposed model. The source code is available at: https://github.com/LeongDong/S2DNets}{https://github.com/LeongDong/S2DNets.
* 11 pages, 3 figures, accepted by MICCAI
Finding Local Diffusion Schrödinger Bridge using Kolmogorov-Arnold Network
Feb 27, 2025Abstract:In image generation, Schr\"odinger Bridge (SB)-based methods theoretically enhance the efficiency and quality compared to the diffusion models by finding the least costly path between two distributions. However, they are computationally expensive and time-consuming when applied to complex image data. The reason is that they focus on fitting globally optimal paths in high-dimensional spaces, directly generating images as next step on the path using complex networks through self-supervised training, which typically results in a gap with the global optimum. Meanwhile, most diffusion models are in the same path subspace generated by weights $f_A(t)$ and $f_B(t)$, as they follow the paradigm ($x_t = f_A(t)x_{Img} + f_B(t)\epsilon$). To address the limitations of SB-based methods, this paper proposes for the first time to find local Diffusion Schr\"odinger Bridges (LDSB) in the diffusion path subspace, which strengthens the connection between the SB problem and diffusion models. Specifically, our method optimizes the diffusion paths using Kolmogorov-Arnold Network (KAN), which has the advantage of resistance to forgetting and continuous output. The experiment shows that our LDSB significantly improves the quality and efficiency of image generation using the same pre-trained denoising network and the KAN for optimising is only less than 0.1MB. The FID metric is reduced by \textbf{more than 15\%}, especially with a reduction of 48.50\% when NFE of DDIM is $5$ for the CelebA dataset. Code is available at https://github.com/Qiu-XY/LDSB.
Tumor Detection, Segmentation and Classification Challenge on Automated 3D Breast Ultrasound: The TDSC-ABUS Challenge
Jan 26, 2025



Abstract:Breast cancer is one of the most common causes of death among women worldwide. Early detection helps in reducing the number of deaths. Automated 3D Breast Ultrasound (ABUS) is a newer approach for breast screening, which has many advantages over handheld mammography such as safety, speed, and higher detection rate of breast cancer. Tumor detection, segmentation, and classification are key components in the analysis of medical images, especially challenging in the context of 3D ABUS due to the significant variability in tumor size and shape, unclear tumor boundaries, and a low signal-to-noise ratio. The lack of publicly accessible, well-labeled ABUS datasets further hinders the advancement of systems for breast tumor analysis. Addressing this gap, we have organized the inaugural Tumor Detection, Segmentation, and Classification Challenge on Automated 3D Breast Ultrasound 2023 (TDSC-ABUS2023). This initiative aims to spearhead research in this field and create a definitive benchmark for tasks associated with 3D ABUS image analysis. In this paper, we summarize the top-performing algorithms from the challenge and provide critical analysis for ABUS image examination. We offer the TDSC-ABUS challenge as an open-access platform at https://tdsc-abus2023.grand-challenge.org/ to benchmark and inspire future developments in algorithmic research.
MedFILIP: Medical Fine-grained Language-Image Pre-training
Jan 18, 2025Abstract:Medical vision-language pretraining (VLP) that leverages naturally-paired medical image-report data is crucial for medical image analysis. However, existing methods struggle to accurately characterize associations between images and diseases, leading to inaccurate or incomplete diagnostic results. In this work, we propose MedFILIP, a fine-grained VLP model, introduces medical image-specific knowledge through contrastive learning, specifically: 1) An information extractor based on a large language model is proposed to decouple comprehensive disease details from reports, which excels in extracting disease deals through flexible prompt engineering, thereby effectively reducing text complexity while retaining rich information at a tiny cost. 2) A knowledge injector is proposed to construct relationships between categories and visual attributes, which help the model to make judgments based on image features, and fosters knowledge extrapolation to unfamiliar disease categories. 3) A semantic similarity matrix based on fine-grained annotations is proposed, providing smoother, information-richer labels, thus allowing fine-grained image-text alignment. 4) We validate MedFILIP on numerous datasets, e.g., RSNA-Pneumonia, NIH ChestX-ray14, VinBigData, and COVID-19. For single-label, multi-label, and fine-grained classification, our model achieves state-of-the-art performance, the classification accuracy has increased by a maximum of 6.69\%. The code is available in https://github.com/PerceptionComputingLab/MedFILIP.
Learning Spatially Decoupled Color Representations for Facial Image Colorization
Dec 10, 2024Abstract:Image colorization methods have shown prominent performance on natural images. However, since humans are more sensitive to faces, existing methods are insufficient to meet the demands when applied to facial images, typically showing unnatural and uneven colorization results. In this paper, we investigate the facial image colorization task and find that the problems with facial images can be attributed to an insufficient understanding of facial components. As a remedy, by introducing facial component priors, we present a novel facial image colorization framework dubbed FCNet. Specifically, we learn a decoupled color representation for each face component (e.g., lips, skin, eyes, and hair) under the guidance of face parsing maps. A chromatic and spatial augmentation strategy is presented to facilitate the learning procedure, which requires only grayscale and color facial image pairs. After training, the presented FCNet can be naturally applied to facial image colorization with single or multiple reference images. To expand the application paradigms to scenarios with no reference images, we further train two alternative modules, which predict the color representations from the grayscale input or a random seed, respectively. Extensive experiments show that our method can perform favorably against existing methods in various application scenarios (i.e., no-, single-, and multi-reference facial image colorization). The source code and pre-trained models will be publicly available.
Unsupervised Decomposition Networks for Bias Field Correction in MR Image
Jul 30, 2023



Abstract:Bias field, which is caused by imperfect MR devices or imaged objects, introduces intensity inhomogeneity into MR images and degrades the performance of MR image analysis methods. Many retrospective algorithms were developed to facilitate the bias correction, to which the deep learning-based methods outperformed. However, in the training phase, the supervised deep learning-based methods heavily rely on the synthesized bias field. As the formation of the bias field is extremely complex, it is difficult to mimic the true physical property of MR images by synthesized data. While bias field correction and image segmentation are strongly related, the segmentation map is precisely obtained by decoupling the bias field from the original MR image, and the bias value is indicated by the segmentation map in reverse. Thus, we proposed novel unsupervised decomposition networks that are trained only with biased data to obtain the bias-free MR images. Networks are made up of: a segmentation part to predict the probability of every pixel belonging to each class, and an estimation part to calculate the bias field, which are optimized alternately. Furthermore, loss functions based on the combination of fuzzy clustering and the multiplicative bias field are also devised. The proposed loss functions introduce the smoothness of bias field and construct the soft relationships among different classes under intra-consistency constraints. Extensive experiments demonstrate that the proposed method can accurately estimate bias fields and produce better bias correction results. The code is available on the link: https://github.com/LeongDong/Bias-Decomposition-Networks.
Efficient automatic segmentation for multi-level pulmonary arteries: The PARSE challenge
Apr 07, 2023



Abstract:Efficient automatic segmentation of multi-level (i.e. main and branch) pulmonary arteries (PA) in CTPA images plays a significant role in clinical applications. However, most existing methods concentrate only on main PA or branch PA segmentation separately and ignore segmentation efficiency. Besides, there is no public large-scale dataset focused on PA segmentation, which makes it highly challenging to compare the different methods. To benchmark multi-level PA segmentation algorithms, we organized the first \textbf{P}ulmonary \textbf{AR}tery \textbf{SE}gmentation (PARSE) challenge. On the one hand, we focus on both the main PA and the branch PA segmentation. On the other hand, for better clinical application, we assign the same score weight to segmentation efficiency (mainly running time and GPU memory consumption during inference) while ensuring PA segmentation accuracy. We present a summary of the top algorithms and offer some suggestions for efficient and accurate multi-level PA automatic segmentation. We provide the PARSE challenge as open-access for the community to benchmark future algorithm developments at \url{https://parse2022.grand-challenge.org/Parse2022/}.
The state-of-the-art 3D anisotropic intracranial hemorrhage segmentation on non-contrast head CT: The INSTANCE challenge
Jan 12, 2023Abstract:Automatic intracranial hemorrhage segmentation in 3D non-contrast head CT (NCCT) scans is significant in clinical practice. Existing hemorrhage segmentation methods usually ignores the anisotropic nature of the NCCT, and are evaluated on different in-house datasets with distinct metrics, making it highly challenging to improve segmentation performance and perform objective comparisons among different methods. The INSTANCE 2022 was a grand challenge held in conjunction with the 2022 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). It is intended to resolve the above-mentioned problems and promote the development of both intracranial hemorrhage segmentation and anisotropic data processing. The INSTANCE released a training set of 100 cases with ground-truth and a validation set with 30 cases without ground-truth labels that were available to the participants. A held-out testing set with 70 cases is utilized for the final evaluation and ranking. The methods from different participants are ranked based on four metrics, including Dice Similarity Coefficient (DSC), Hausdorff Distance (HD), Relative Volume Difference (RVD) and Normalized Surface Dice (NSD). A total of 13 teams submitted distinct solutions to resolve the challenges, making several baseline models, pre-processing strategies and anisotropic data processing techniques available to future researchers. The winner method achieved an average DSC of 0.6925, demonstrating a significant growth over our proposed baseline method. To the best of our knowledge, the proposed INSTANCE challenge releases the first intracranial hemorrhage segmentation benchmark, and is also the first challenge that intended to resolve the anisotropic problem in 3D medical image segmentation, which provides new alternatives in these research fields.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge