Chulhong Kim
GPAIR: Gaussian-Kernel-Based Ultrafast 3D Photoacoustic Iterative Reconstruction
Feb 03, 2026Abstract:Although the iterative reconstruction (IR) algorithm can substantially correct reconstruction artifacts in photoacoustic (PA) computed tomography (PACT), it suffers from long reconstruction times, especially for large-scale three-dimensional (3D) imaging in which IR takes hundreds of seconds to hours. The computing burden severely limits the practical applicability of IR algorithms. In this work, we proposed an ultrafast IR method for 3D PACT, called Gaussian-kernel-based Ultrafast 3D Photoacoustic Iterative Reconstruction (GPAIR), which achieves orders-of-magnitude acceleration in computing. GPAIR transforms traditional spatial grids with continuous isotropic Gaussian kernels. By deriving analytical closed-form expression for pressure waves and implementing powerful GPU-accelerated differentiable Triton operators, GPAIR demonstrates extraordinary ultrafast sub-second reconstruction speed for 3D targets containing 8.4 million voxels in animal experiments. This revolutionary ultrafast image reconstruction enables near-real-time large-scale 3D PA reconstruction, significantly advancing 3D PACT toward clinical applications.
SlingBAG Pro: Accelerating point cloud-based iterative reconstruction for 3D photoacoustic imaging with arbitrary array geometries
Jan 06, 2026Abstract:High-quality three-dimensional (3D) photoacoustic imaging (PAI) is gaining increasing attention in clinical applications. To address the challenges of limited space and high costs, irregular geometric transducer arrays that conform to specific imaging regions are promising for achieving high-quality 3D PAI with fewer transducers. However, traditional iterative reconstruction algorithms struggle with irregular array configurations, suffering from high computational complexity, substantial memory requirements, and lengthy reconstruction times. In this work, we introduce SlingBAG Pro, an advanced reconstruction algorithm based on the point cloud iteration concept of the Sliding ball adaptive growth (SlingBAG) method, while extending its compatibility to arbitrary array geometries. SlingBAG Pro maintains high reconstruction quality, reduces the number of required transducers, and employs a hierarchical optimization strategy that combines zero-gradient filtering with progressively increased temporal sampling rates during iteration. This strategy rapidly removes redundant spatial point clouds, accelerates convergence, and significantly shortens overall reconstruction time. Compared to the original SlingBAG algorithm, SlingBAG Pro achieves up to a 2.2-fold speed improvement in point cloud-based 3D PA reconstruction under irregular array geometries. The proposed method is validated through both simulation and in vivo mouse experiments, and the source code is publicly available at https://github.com/JaegerCQ/SlingBAG_Pro.
Tumor Detection, Segmentation and Classification Challenge on Automated 3D Breast Ultrasound: The TDSC-ABUS Challenge
Jan 26, 2025



Abstract:Breast cancer is one of the most common causes of death among women worldwide. Early detection helps in reducing the number of deaths. Automated 3D Breast Ultrasound (ABUS) is a newer approach for breast screening, which has many advantages over handheld mammography such as safety, speed, and higher detection rate of breast cancer. Tumor detection, segmentation, and classification are key components in the analysis of medical images, especially challenging in the context of 3D ABUS due to the significant variability in tumor size and shape, unclear tumor boundaries, and a low signal-to-noise ratio. The lack of publicly accessible, well-labeled ABUS datasets further hinders the advancement of systems for breast tumor analysis. Addressing this gap, we have organized the inaugural Tumor Detection, Segmentation, and Classification Challenge on Automated 3D Breast Ultrasound 2023 (TDSC-ABUS2023). This initiative aims to spearhead research in this field and create a definitive benchmark for tasks associated with 3D ABUS image analysis. In this paper, we summarize the top-performing algorithms from the challenge and provide critical analysis for ABUS image examination. We offer the TDSC-ABUS challenge as an open-access platform at https://tdsc-abus2023.grand-challenge.org/ to benchmark and inspire future developments in algorithmic research.
Zero-Shot Artifact2Artifact: Self-incentive artifact removal for photoacoustic imaging without any data
Dec 19, 2024



Abstract:Photoacoustic imaging (PAI) uniquely combines optical contrast with the penetration depth of ultrasound, making it critical for clinical applications. However, the quality of 3D PAI is often degraded due to reconstruction artifacts caused by the sparse and angle-limited configuration of detector arrays. Existing iterative or deep learning-based methods are either time-consuming or require large training datasets, significantly limiting their practical application. Here, we propose Zero-Shot Artifact2Artifact (ZS-A2A), a zero-shot self-supervised artifact removal method based on a super-lightweight network, which leverages the fact that reconstruction artifacts are sensitive to irregularities caused by data loss. By introducing random perturbations to the acquired PA data, it spontaneously generates subset data, which in turn stimulates the network to learn the artifact patterns in the reconstruction results, thus enabling zero-shot artifact removal. This approach requires neither training data nor prior knowledge of the artifacts, and is capable of artifact removal for 3D PAI. For maximum amplitude projection (MAP) images or slice images in 3D PAI acquired with arbitrarily sparse or angle-limited detector arrays, ZS-A2A employs a self-incentive strategy to complete artifact removal and improves the Contrast-to-Noise Ratio (CNR). We validated ZS-A2A in both simulation study and $ in\ vivo $ animal experiments. Results demonstrate that ZS-A2A achieves state-of-the-art (SOTA) performance compared to existing zero-shot methods, and for the $ in\ vivo $ rat liver, ZS-A2A improves CNR from 17.48 to 43.46 in just 8 seconds. The project for ZS-A2A will be available in the following GitHub repository: https://github.com/JaegerCQ/ZS-A2A.
4D SlingBAG: spatial-temporal coupled Gaussian ball for large-scale dynamic 3D photoacoustic iterative reconstruction
Dec 05, 2024Abstract:Large-scale dynamic three-dimensional (3D) photoacoustic imaging (PAI) is significantly important in clinical applications. In practical implementations, large-scale 3D real-time PAI systems typically utilize sparse two-dimensional (2D) sensor arrays with certain angular deficiencies, necessitating advanced iterative reconstruction (IR) algorithms to achieve quantitative PAI and reduce reconstruction artifacts. However, for existing IR algorithms, multi-frame 3D reconstruction leads to extremely high memory consumption and prolonged computation time, with limited consideration of the spatial-temporal continuity between data frames. Here, we propose a novel method, named the 4D sliding Gaussian ball adaptive growth (4D SlingBAG) algorithm, based on the current point cloud-based IR algorithm sliding Gaussian ball adaptive growth (SlingBAG), which has minimal memory consumption among IR methods. Our 4D SlingBAG method applies spatial-temporal coupled deformation functions to each Gaussian sphere in point cloud, thus explicitly learning the deformations features of the dynamic 3D PA scene. This allows for the efficient representation of various physiological processes (such as pulsation) or external pressures (e.g., blood perfusion experiments) contributing to changes in vessel morphology and blood flow during dynamic 3D PAI, enabling highly efficient IR for dynamic 3D PAI. Simulation experiments demonstrate that 4D SlingBAG achieves high-quality dynamic 3D PA reconstruction. Compared to performing reconstructions by using SlingBAG algorithm individually for each frame, our method significantly reduces computational time and keeps a extremely low memory consumption. The project for 4D SlingBAG can be found in the following GitHub repository: \href{https://github.com/JaegerCQ/4D-SlingBAG}{https://github.com/JaegerCQ/4D-SlingBAG}.
SlingBAG: Sliding ball adaptive growth algorithm with differentiable radiation enables super-efficient iterative 3D photoacoustic image reconstruction
Jul 16, 2024



Abstract:High-quality 3D photoacoustic imaging (PAI) reconstruction under sparse view or limited view has long been challenging. Traditional 3D iterative-based reconstruction methods suffer from both slow speed and high memory consumption. Recently, in computer graphics, the differentiable rendering has made significant progress, particularly with the rise of 3D Gaussian Splatting. Inspired by these, we introduce differentiable radiation into PAI, developing a novel reconstruction algorithm: the Sliding Ball Adaptive Growth algorithm (SlingBAG) for 3D PAI, which shows ability in high-quality 3D PAI reconstruction both under extremely sparse view and limited view. We established the point cloud dataset in PAI, and used unique differentiable rapid radiator based on the spherical decomposition strategy and the randomly initialized point cloud adaptively optimized according to sparse sensor data. Each point undergoes updates in 3D coordinates, initial pressure, and resolution (denoted by the radius of ball). Points undergo adaptive growth during iterative process, including point destroying, splitting and duplicating along the gradient of their positions, manifesting the sliding ball effect. Finally, our point cloud to voxel grid shader renders the final reconstruction results. Simulation and in vivo experiments demonstrate that our SlingBAG reconstruction result's SNR can be more than 40 dB under extremely sparse view, while the SNR of traditional back-projection algorithm's result is less than 20 dB. Moreover, the result of SlingBAG's structural similarity to the ground truth is significantly higher, with an SSIM value of 95.6%. Notably, our differentiable rapid radiator can conduct forward PA simulation in homogeneous, non-viscous media substantially faster than current methods that numerically simulate the wave propagation, such as k-Wave. The dataset and all code will be open source.
A Robust Ensemble Algorithm for Ischemic Stroke Lesion Segmentation: Generalizability and Clinical Utility Beyond the ISLES Challenge
Apr 03, 2024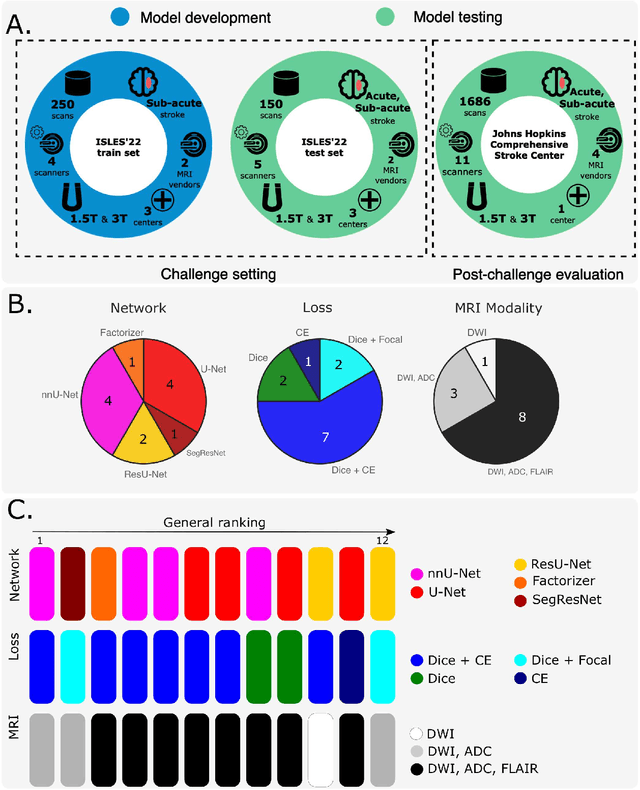
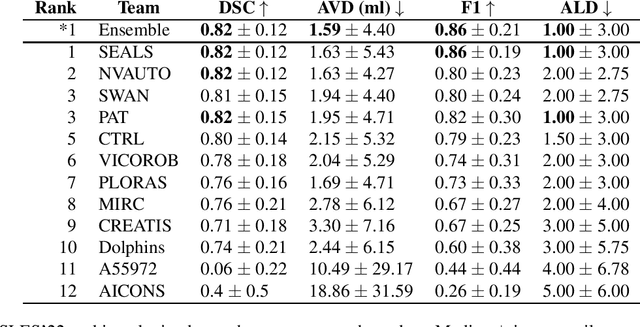
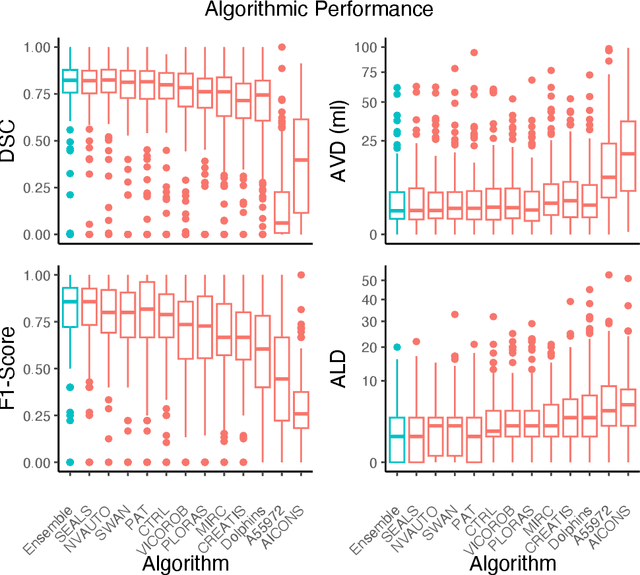
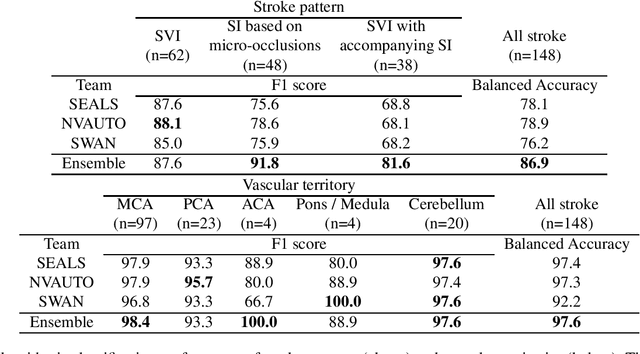
Abstract:Diffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (https://github.com/Tabrisrei/ISLES22_Ensemble) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
Ensemble Transfer Learning of Elastography and B-mode Breast Ultrasound Images
Feb 17, 2021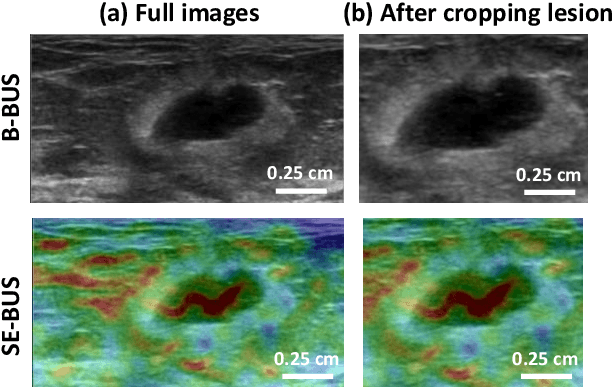
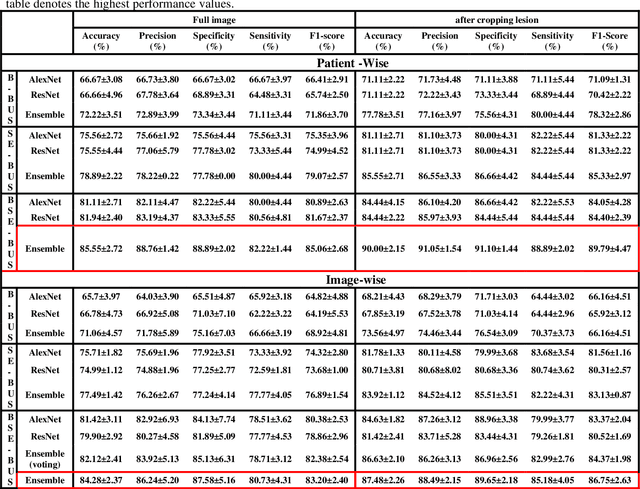
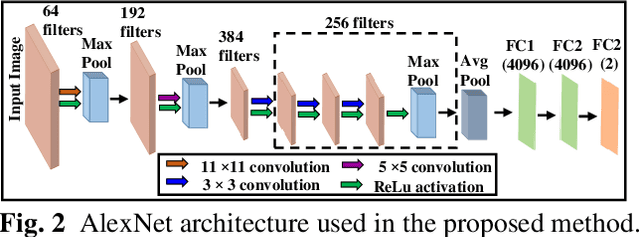
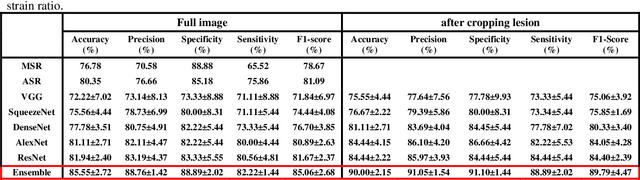
Abstract:Computer-aided detection (CAD) of benign and malignant breast lesions becomes increasingly essential in breast ultrasound (US) imaging. The CAD systems rely on imaging features identified by the medical experts for their performance, whereas deep learning (DL) methods automatically extract features from the data. The challenge of the DL is the insufficiency of breast US images available to train the DL models. Here, we present an ensemble transfer learning model to classify benign and malignant breast tumors using B-mode breast US (B-US) and strain elastography breast US (SE-US) images. This model combines semantic features from AlexNet & ResNet models to classify benign from malignant tumors. We use both B-US and SE-US images to train the model and classify the tumors. We retrospectively gathered 85 patients' data, with 42 benign and 43 malignant cases confirmed with the biopsy. Each patient had multiple B-US and their corresponding SE-US images, and the total dataset contained 261 B-US images and 261 SE-US images. Experimental results show that our ensemble model achieves a sensitivity of 88.89% and specificity of 91.10%. These diagnostic performances of the proposed method are equivalent to or better than manual identification. Thus, our proposed ensemble learning method would facilitate detecting early breast cancer, reliably improving patient care.
Multi-Channel Transfer Learning of Chest X-ray Images for Screening of COVID-19
May 12, 2020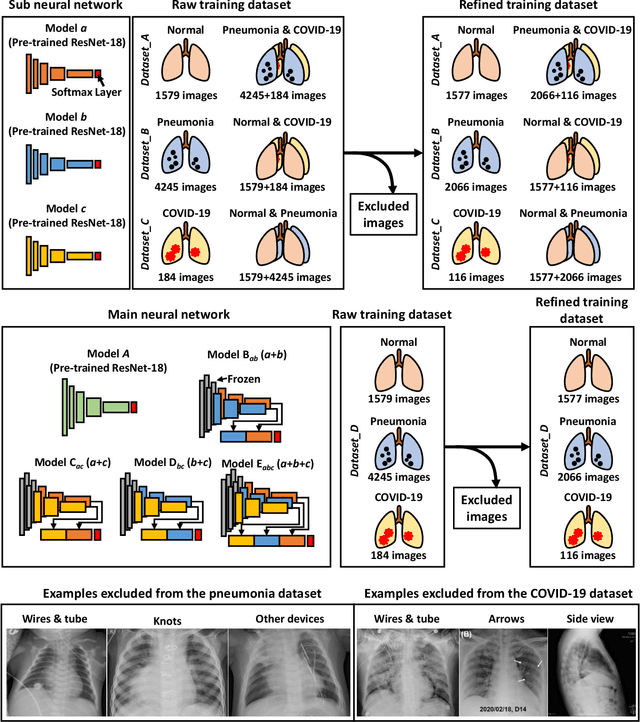
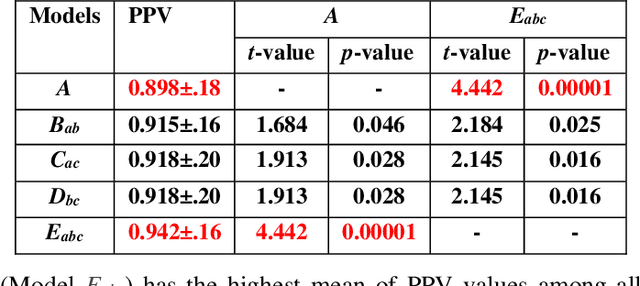
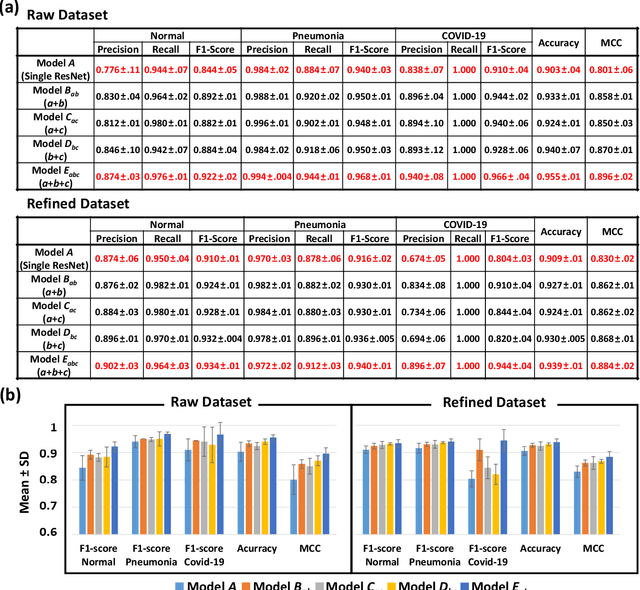
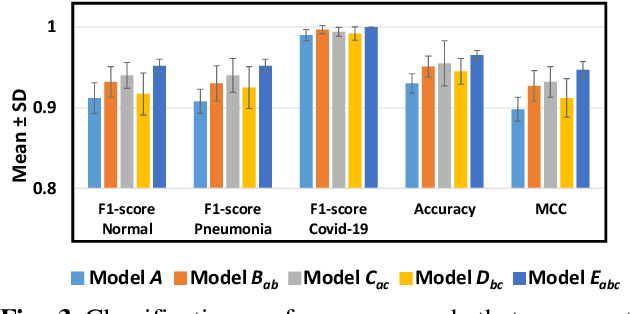
Abstract:The 2019 novel coronavirus (COVID-19) has spread rapidly all over the world and it is affecting the whole society. The current gold standard test for screening COVID-19 patients is the polymerase chain reaction test. However, the COVID-19 test kits are not widely available and time-consuming. Thus, as an alternative, chest X-rays are being considered for quick screening. Since the presentation of COVID-19 in chest X-rays is varied in features and specialization in reading COVID-19 chest X-rays are required thus limiting its use for diagnosis. To address this challenge of reading chest X-rays by radiologists quickly, we present a multi-channel transfer learning model based on ResNet architecture to facilitate the diagnosis of COVID-19 chest X-ray. Three ResNet-based models (Models a, b, and c) were retrained using Dataset_A (1579 normal and 4429 diseased), Dataset_B (4245 pneumonia and 1763 non-pneumonia), and Dataset_C (184 COVID-19 and 5824 Non-COVID19), respectively, to classify (a) normal or diseased, (b) pneumonia or non-pneumonia, and (c) COVID-19 or non-COVID19. Finally, these three models were ensembled and fine-tuned using Dataset_D (1579 normal, 4245 pneumonia, and 184 COVID-19) to classify normal, pneumonia, and COVID-19 cases. Our results show that the ensemble model is more accurate than the single ResNet model, which is also re-trained using Dataset_D as it extracts more relevant semantic features for each class. Our approach provides a precision of 94 % and a recall of 100%. Thus, our method could potentially help clinicians in screening patients for COVID-19, thus facilitating immediate triaging and treatment for better outcomes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge